CN:Tak1 complex explains CN substrate specificity
Shirakawa, K.T., Parikh, T., Page, R., Peti, W.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Protein phosphatase 3 catalytic subunit alpha | 370 | Homo sapiens | Mutation(s): 1 Gene Names: PPP3CA, CALNA, CNA EC: 3.1.3.16 | 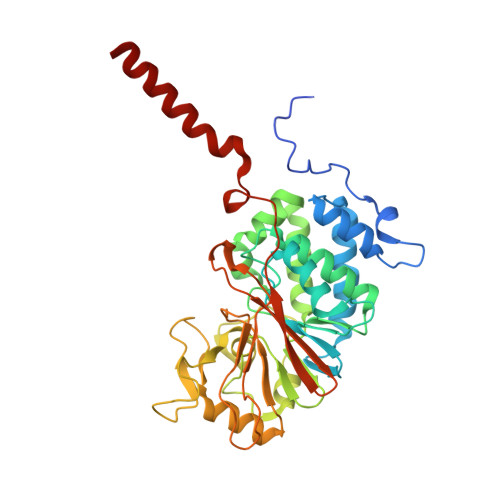 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q08209 (Homo sapiens) Explore Q08209 Go to UniProtKB: Q08209 | |||||
PHAROS: Q08209 GTEx: ENSG00000138814 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q08209 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Calcineurin subunit B type 1 | 156 | Homo sapiens | Mutation(s): 0 Gene Names: PPP3R1, CNA2, CNB | 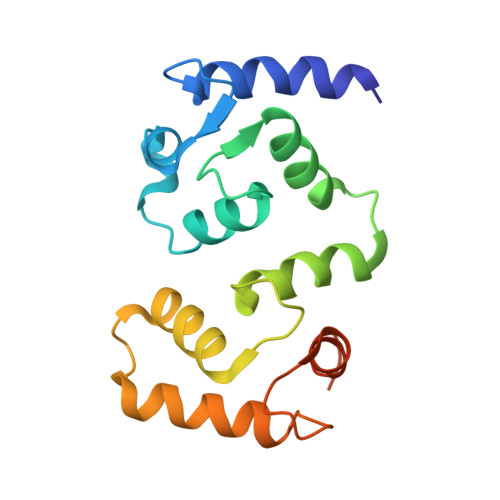 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P63098 (Homo sapiens) Explore P63098 Go to UniProtKB: P63098 | |||||
PHAROS: P63098 GTEx: ENSG00000221823 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P63098 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| IkB-like protein,Mitogen-activated protein kinase kinase kinase 7 | 53 | Homo sapiens | Mutation(s): 2 Gene Names: A238L, Mal-047, 5EL, MAP3K7, TAK1 EC: 2.7.11.25 | 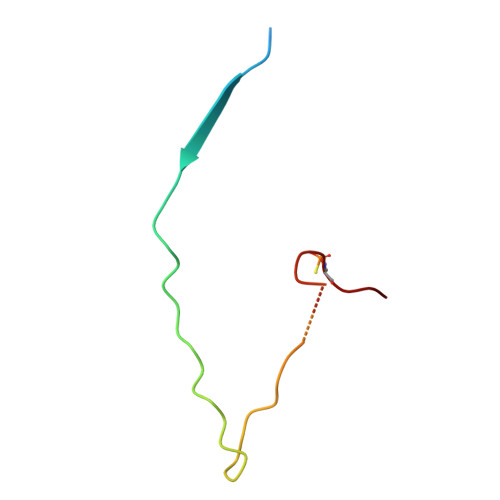 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for O36972 (African swine fever virus (isolate Tick/Malawi/Lil 20-1/1983)) Explore O36972 Go to UniProtKB: O36972 | |||||
Find proteins for O43318 (Homo sapiens) Explore O43318 Go to UniProtKB: O43318 | |||||
PHAROS: O43318 GTEx: ENSG00000135341 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Groups | O43318O36972 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 4 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| PEG Query on PEG | H [auth A], M [auth B], P [auth D] | DI(HYDROXYETHYL)ETHER C4 H10 O3 MTHSVFCYNBDYFN-UHFFFAOYSA-N |  | ||
| GOL Query on GOL | O [auth D] | GLYCEROL C3 H8 O3 PEDCQBHIVMGVHV-UHFFFAOYSA-N |  | ||
| FE Query on FE | G [auth A], N [auth D] | FE (III) ION Fe VTLYFUHAOXGGBS-UHFFFAOYSA-N |  | ||
| CA Query on CA | I [auth B] J [auth B] K [auth B] L [auth B] Q [auth E] | CALCIUM ION Ca BHPQYMZQTOCNFJ-UHFFFAOYSA-N |  | ||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| 2RX Query on 2RX | C, F | L-PEPTIDE LINKING | C3 H8 N O5 P S |  | SER |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 224.079 | α = 90 |
| b = 49.585 | β = 128.72 |
| c = 158.895 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| autoPROC | data reduction |
| autoPROC | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) | United States | R01GM144379 |
| National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) | United States | R01NS124666 |