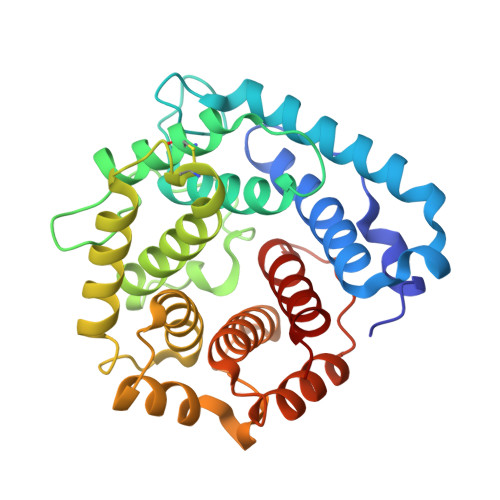
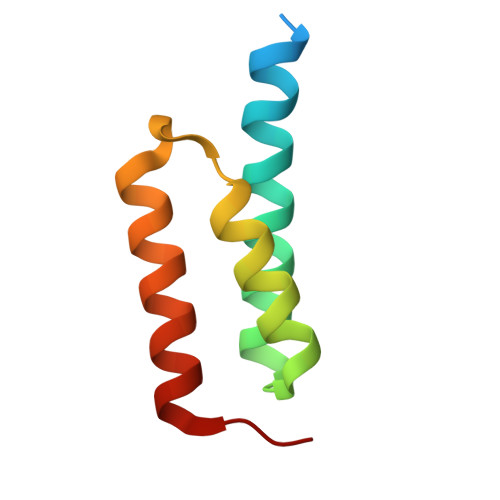
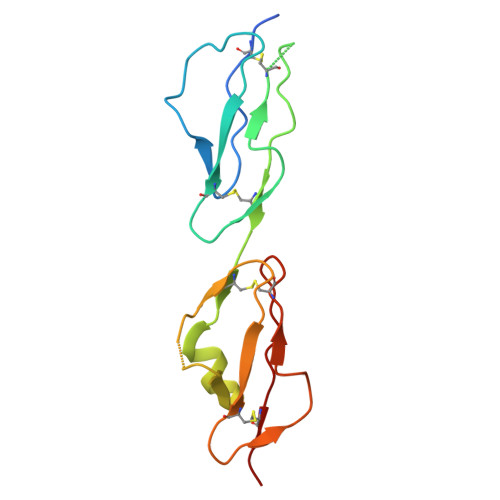
The C-terminal domain of Staphylococcus aureus Efb recruits FHR-2 to C3b, synergistically inhibiting the terminal complement pathway.
Duan, H., Kortvely, E., Mary, J.L., Hauck, S.M., Geisbrecht, B.V.(2025) J Immunol
- PubMed: 41273729
- DOI: https://doi.org/10.1093/jimmun/vkaf316
- Primary Citation of Related Structures:
9N1Z, 9N20 - PubMed Abstract:
The extracellular fibrinogen-binding protein (Efb) is one of nearly a dozen proteins secreted by Staphylococcus aureus to inhibit complement activation or amplification. The C-terminal domain of Efb (Efb-C) forms a high-affinity interaction with the thioester-containing domain of C3b (TED/C3d), thereby blocking formation of the C3 proconvertase complex through an allosteric mechanism. However, further functional consequences of Efb-C binding to C3b remain unexplored. Here, we identified a previously unknown interaction between Efb-C, C3b, and the complement regulatory molecule FHR2 (factor H-related protein 2). Since the FHR2/C3b interaction is centered upon the 2 C-terminal-most domains of FHR2 (FHR2[3-4]) and the TED/C3d domain of C3b, we tested whether Efb-C could influence the FHR2(3-4)/C3d interaction. We observed a significant enhancement of FHR2(3-4)/C3d binding in the presence of Efb-C. We studied the FHR2(3-4)/C3d/Efb-C complex by X-ray crystallography and found that Efb-C forms few direct interactions with FHR2(3-4). Yet, the presence of Efb-C also enhanced binding of FHR2(3-4) and full-length FHR2 to C3b, suggesting that the effect of Efb-C on the FHR2/C3b interaction arises from increased accessibility of the FHR2-binding site. We found that enhanced FHR2 binding did not impact the rate of C3 convertase formation more than Efb-C alone, nor did it impart decay acceleration or cofactor activity. However, we observed potent, synergistic inhibition of complement downstream of C5 activation by Efb-C and FHR2 but not by Efb-C and FHR2(3-4). Our results show that Efb-C binding to C3b exerts additional inhibitory effects on the central complement components beyond blocking formation of the C3 proconvertase alone.
- Department of Biochemistry and Molecular Biophysics, Kansas State University, Manhattan, KS, United States.
Organizational Affiliation: