The crystal structure of SARS-CoV-2 NSP5 in complex with PTBP1
Yan, X., Jin, Z., Wang, Y., Zhang, J.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 3C-like proteinase nsp5 | 306 | Severe acute respiratory syndrome coronavirus 2 | Mutation(s): 0 Gene Names: rep, 1a-1b EC: 3.4.22.69 | 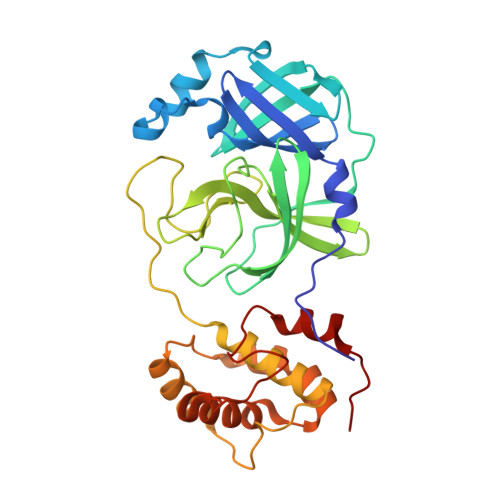 | |
UniProt | |||||
Find proteins for P0DTD1 (Severe acute respiratory syndrome coronavirus 2) Explore P0DTD1 Go to UniProtKB: P0DTD1 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0DTD1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by: Sequence | 3D Structure
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Polypyrimidine tract-binding protein 1 | B [auth C] | 10 | Homo sapiens | Mutation(s): 0 | 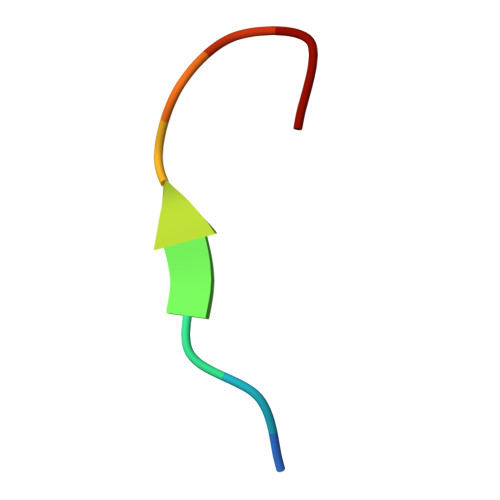 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P26599 (Homo sapiens) Explore P26599 Go to UniProtKB: P26599 | |||||
PHAROS: P26599 GTEx: ENSG00000011304 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P26599 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 96.154 | α = 90 |
| b = 81.651 | β = 116.99 |
| c = 54.334 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| XDS | data scaling |
| XDS | data reduction |
| PHENIX | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Natural Science Foundation of China (NSFC) | China | -- |