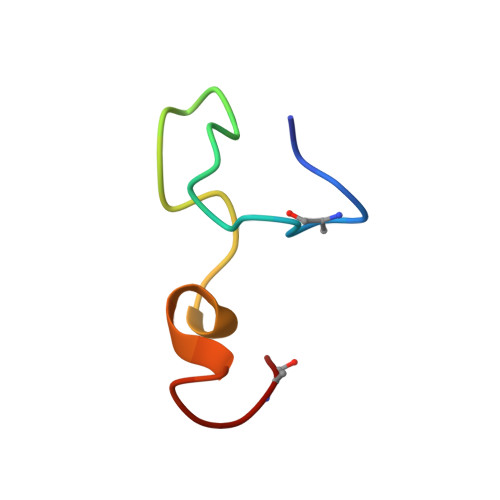
Isolation, Characterization and Structure Elucidation of a Novel Lantibiotic From Paenibacillus sp.
Karczewski, J., Krasucki, S.P., Asare-Okai, P.N., Diehl, C., Friedman, A., Brown, C.M., Maezato, Y., Streatfield, S.J.(2020) Front Microbiol 11: 598789-598789
- PubMed: 33324379
- DOI: https://doi.org/10.3389/fmicb.2020.598789
- Primary Citation of Related Structures:
7K1Q - PubMed Abstract:
We have isolated and characterized a novel antibacterial peptide, CMB001, following an extensive screening effort of bacterial species isolated from diverse environmental sources. The bacterium that produces CMB001 is characterized as a Gram (+) bacillus sharing approximately 98.9% 16S rRNA sequence homology with its closest match, Paenibacillus kyungheensis . The molecule has been purified to homogeneity from its cell-free supernatant by a three-step preparative chromatography process. Based on its primary structure, CMB001 shares 81% identity with subtilin and 62% with nisin. CMB001 is active mainly against Gram-positive bacteria and Mycobacteriaceae but it is also active against certain Gram-negative bacteria, including multi-drug resistant Acinetobacter baumannii . It retains full antibacterial activity at neutral pH and displays a low propensity to select for resistance among targeted bacteria. Based on NMR and mass spectrometry, CMB001 forms a unique 3D-structure comprising of a compact backbone with one α-helix and two pseudo-α-helical regions. Screening the structure against the Protein Data Bank (PDB) revealed a partial match with nisin-lipid II (1WCO), but none of the lantibiotics with known structures showed significant structural similarity. Due to its unique structure, resistance profile, relatively broad spectrum and stability under physiological conditions, CMB001 is a promising drug candidate for evaluation in animal models of bacterial infection.
Organizational Affiliation:
Fraunhofer USA Center for Molecular Biotechnology, Newark, DE, United States.