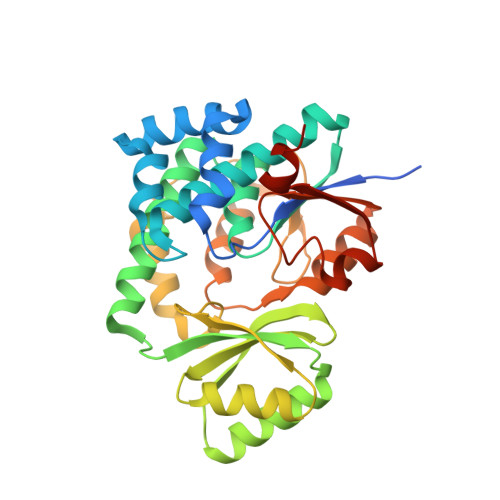
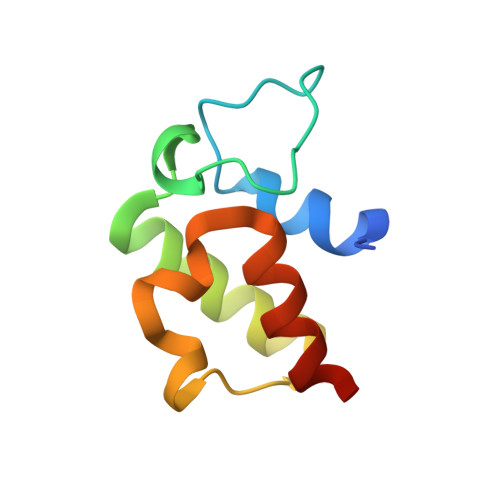
Complex structure of the acyltransferase VinK and the carrier protein VinL with a pantetheine cross-linking probe.
Miyanaga, A., Ouchi, R., Kudo, F., Eguchi, T.(2021) Acta Crystallogr F Struct Biol Commun 77: 294-302
- PubMed: 34473106
- DOI: https://doi.org/10.1107/S2053230X21008761
- Primary Citation of Related Structures:
7F2R - PubMed Abstract:
Acyltransferases are responsible for the selection and loading of acyl units onto carrier proteins in polyketide and fatty-acid biosynthesis. Despite the importance of protein-protein interactions between the acyltransferase and the carrier protein, structural information on acyltransferase-carrier protein interactions is limited because of the transient interactions between them. In the biosynthesis of the polyketide vicenistatin, the acyltransferase VinK recognizes the carrier protein VinL for the transfer of a dipeptidyl unit. The crystal structure of a VinK-VinL covalent complex formed with a 1,2-bismaleimidoethane cross-linking reagent has been determined previously. Here, the crystal structure of a VinK-VinL covalent complex formed with a pantetheine cross-linking probe is reported at 1.95 Å resolution. In the structure of the VinK-VinL-probe complex, the pantetheine probe that is attached to VinL is covalently connected to the side chain of the mutated Cys106 of VinK. The interaction interface between VinK and VinL is essentially the same in the two VinK-VinL complex structures, although the position of the pantetheine linker slightly differs. This structural observation suggests that interface interactions are not affected by the cross-linking strategy used.
Organizational Affiliation:
Department of Chemistry, Tokyo Institute of Technology, 2-12-1 O-okayama, Meguro-ku, Tokyo 152-8551, Japan.