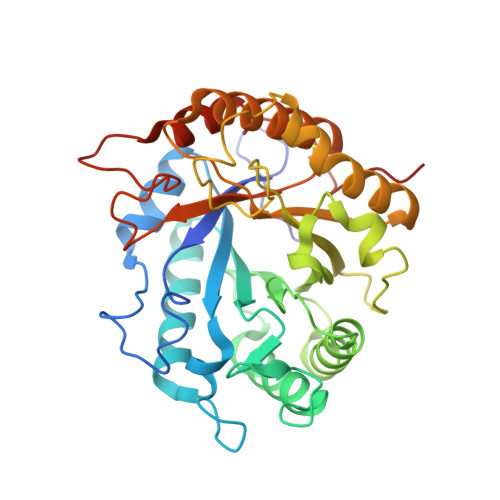
Crystal structure determination of Pseudomonas stutzeri A1501 endoglucanase Cel5A: the search for a molecular basis for glycosynthesis in GH5_5 enzymes.
Dutoit, R., Delsaute, M., Collet, L., Vander Wauven, C., Van Elder, D., Berlemont, R., Richel, A., Galleni, M., Bauvois, C.(2019) Acta Crystallogr D Struct Biol 75: 605-615