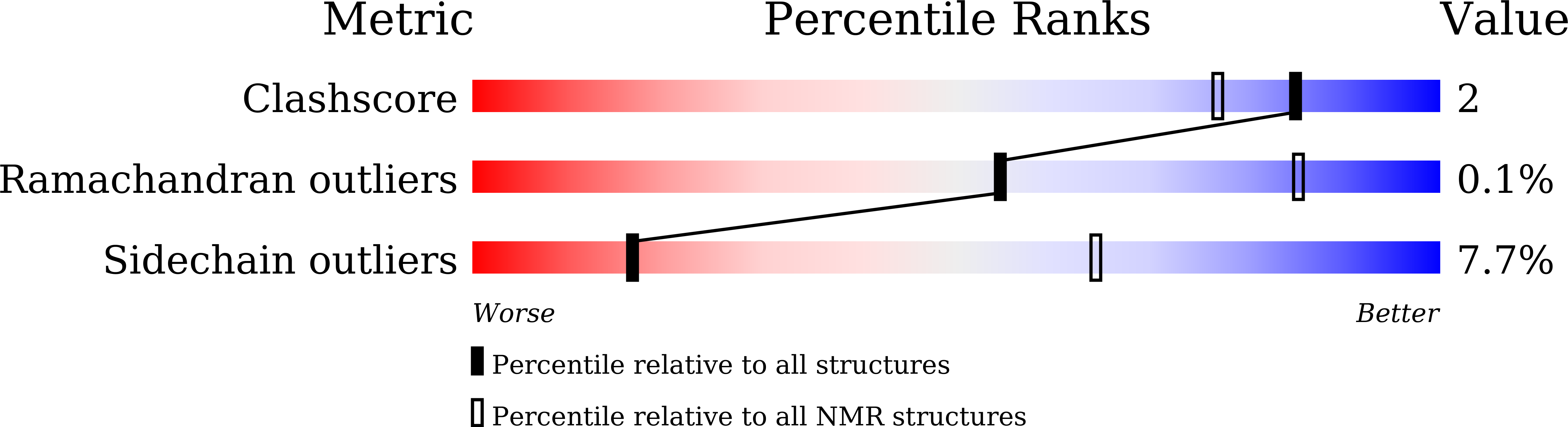Taf14 recognizes a common motif in transcriptional machineries and facilitates their clustering by phase separation.
Chen, G., Wang, D., Wu, B., Yan, F., Xue, H., Wang, Q., Quan, S., Chen, Y.(2020) Nat Commun 11: 4206-4206
- PubMed: 32826896
- DOI: https://doi.org/10.1038/s41467-020-18021-7
- Primary Citation of Related Structures:
6LQZ - PubMed Abstract:
Saccharomyces cerevisiae TBP associated factor 14 (Taf14) is a well-studied transcriptional regulator that controls diverse physiological processes and that physically interacts with at least seven nuclear complexes in yeast. Despite multiple previous Taf14 structural studies, the nature of its disparate transcriptional regulatory functions remains opaque. Here, we demonstrate that the extra-terminal (ET) domain of Taf14 (Taf14 ET ) recognizes a common motif in multiple transcriptional coactivator proteins from several nuclear complexes, including RSC, SWI/SNF, INO80, NuA3, TFIID, and TFIIF. Moreover, we show that such partner binding promotes liquid-liquid phase separation (LLPS) of Taf14 ET , in a mechanism common to YEATS-associated ET domains (e.g., AF9 ET ) but not Bromo-associated ET domains from BET-family proteins. Thus, beyond identifying the molecular mechanism by which Taf14 ET associates with many transcriptional regulators, our study suggests that Taf14 may function as a versatile nuclear hub that orchestrates transcriptional machineries to spatiotemporally regulate diverse cellular pathways.
Organizational Affiliation:
State Key Laboratory of Molecular Biology, National Center for Protein Science Shanghai, Shanghai Institute of Biochemistry and Cell Biology, Center for Excellence in Molecular Cell Science, Chinese Academy of Sciences, Shanghai, 200031, China.