Turning an Asparaginyl Endopeptidase into a Peptide Ligase
Hemu, X., El Sahili, A., Hu, S., Zhang, X., Serra, A., Goh, B.C., Darwis, D.A., Chen, M.W., Sze, S.K., Liu, C.F., Lescar, J., Tam, J.P.(2020) ACS Catal 10: 8825-8834
Experimental Data Snapshot
(2020) ACS Catal 10: 8825-8834
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Asparaginyl endopeptidase | 502 | Clitoria ternatea | Mutation(s): 2 EC: 3.4.22.34 | 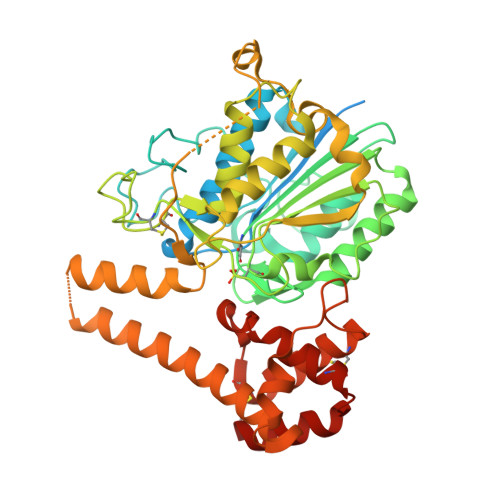 | |
UniProt | |||||
Find proteins for A0A0P0QM28 (Clitoria ternatea) Explore A0A0P0QM28 Go to UniProtKB: A0A0P0QM28 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A0P0QM28 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| NAG Query on NAG | C [auth A], F [auth B] | 2-acetamido-2-deoxy-beta-D-glucopyranose C8 H15 N O6 OVRNDRQMDRJTHS-FMDGEEDCSA-N |  | ||
| PEG Query on PEG | E [auth A] | DI(HYDROXYETHYL)ETHER C4 H10 O3 MTHSVFCYNBDYFN-UHFFFAOYSA-N |  | ||
| EDO Query on EDO | D [auth A] | 1,2-ETHANEDIOL C2 H6 O2 LYCAIKOWRPUZTN-UHFFFAOYSA-N |  | ||
| Modified Residues 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| OCS Query on OCS | A, B | L-PEPTIDE LINKING | C3 H7 N O5 S |  | CYS |
| SNN Query on SNN | A, B | L-PEPTIDE LINKING | C4 H6 N2 O2 |  | ASN |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 44.61 | α = 90 |
| b = 79.6 | β = 93.95 |
| c = 136.47 | γ = 90 |
| Software Name | Purpose |
|---|---|
| BUSTER | refinement |
| XDS | data reduction |
| XDS | data scaling |
| MOLREP | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Ministry of Education (MoE, Singapore) | Singapore | MOE2016-T3-1-003 |