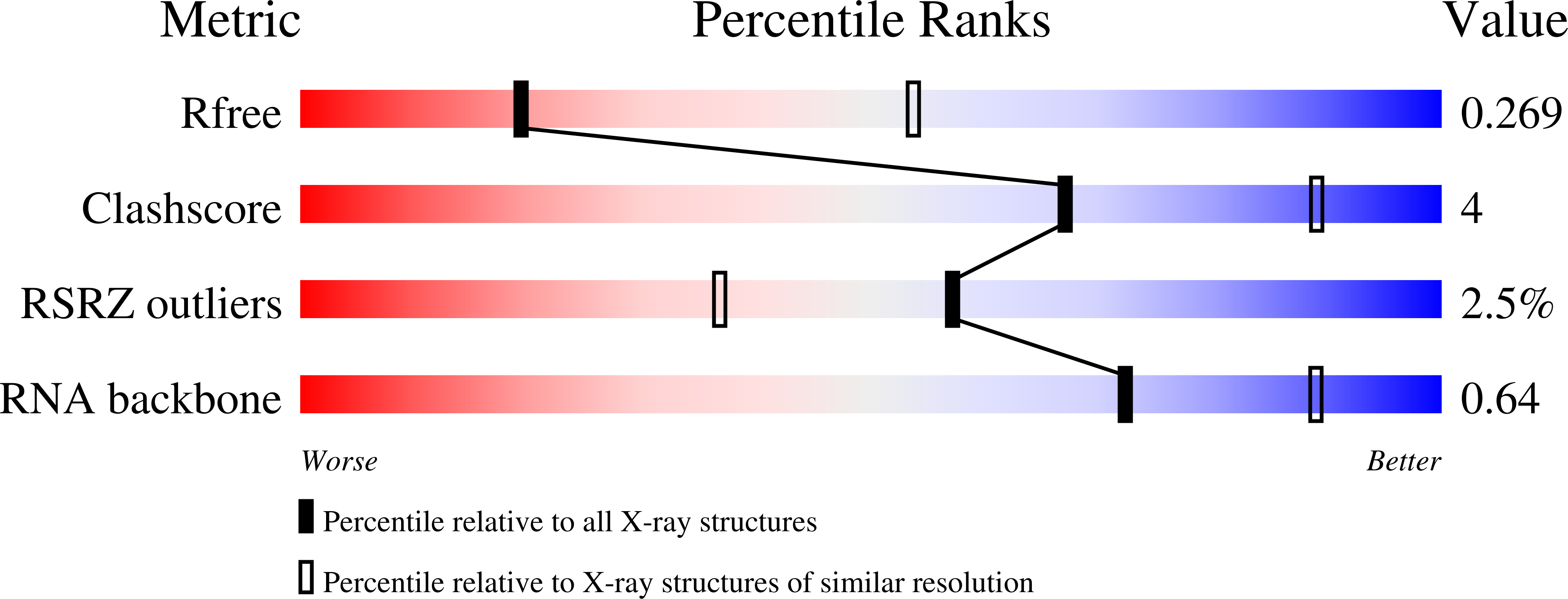
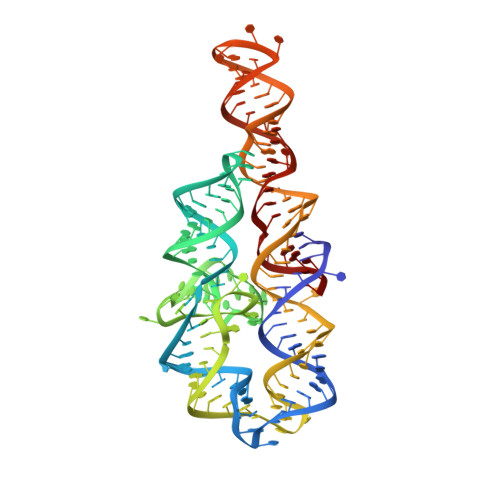
In Crystallo Selection to Establish New RNA Crystal Contacts.
Shoffner, G.M., Wang, R., Podell, E., Cech, T.R., Guo, F.(2018) Structure 26: 1275
- PubMed: 29910185
- DOI: https://doi.org/10.1016/j.str.2018.05.005
- Primary Citation of Related Structures:
6BJX, 6D8L, 6D8M, 6D8N, 6D8O - PubMed Abstract:
Crystallography is a major technique for determining large RNA structures. Obtaining diffraction-quality crystals has been the bottleneck. Although several RNA crystallization methods have been developed, the field strongly needs additional approaches. Here we invented an in crystallo selection strategy for identifying mutations that enhance a target RNA's crystallizability. The strategy includes constructing an RNA pool containing random mutations, obtaining crystals, and amplifying the sequences enriched by crystallization. We demonstrated a proof-of-principle application to the P4-P6 domain from the Tetrahymena ribozyme. We further determined the structures of four selected mutants. All four establish new crystal lattice contacts while maintaining the native structure. Three mutants achieve this by relocating bulges and one by making a helix more flexible. In crystallo selection provides opportunities to improve crystals of RNAs or RNA-ligand complexes. Our results also suggest that mutants may be rationally designed for crystallization by "walking" a bulge along the RNA chain.
Organizational Affiliation:
Department of Biological Chemistry, David Geffen School of Medicine, University of California, Los Angeles, CA 90095, USA.