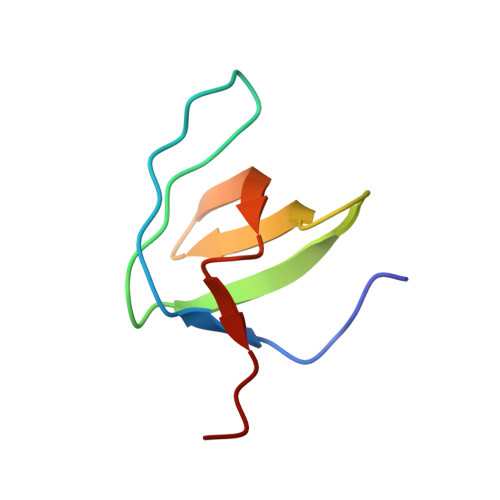
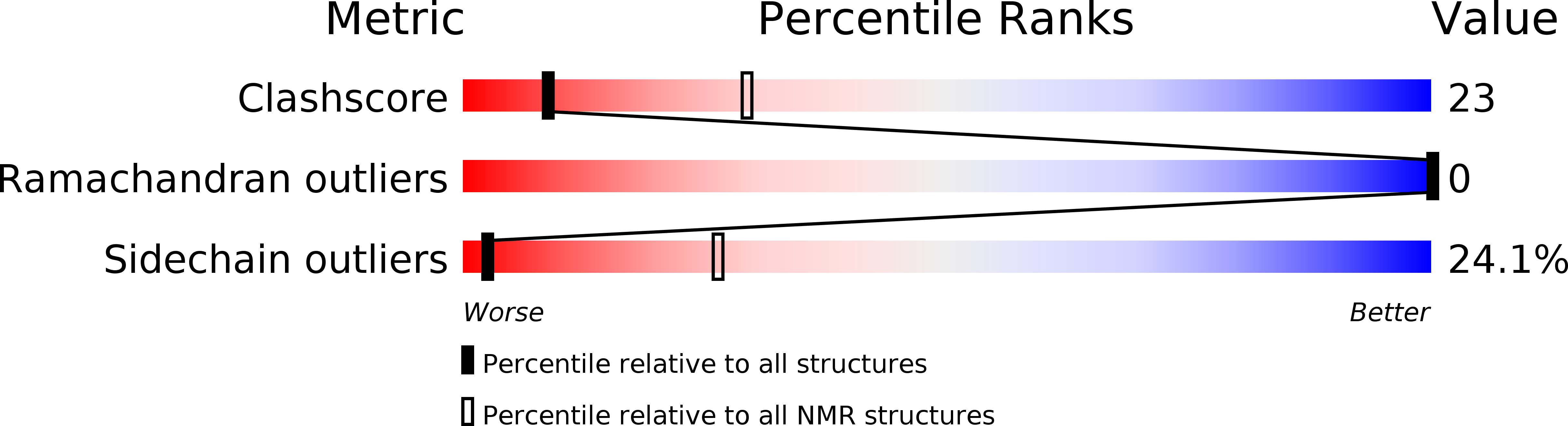Solution structures of the SH3 domains from Shank scaffold proteins and their interactions with Cav1.3 calcium channels.
Ishida, H., Skorobogatov, A., Yamniuk, A.P., Vogel, H.J.(2018) FEBS Lett 592: 2786-2797
- PubMed: 30058071
- DOI: https://doi.org/10.1002/1873-3468.13209
- Primary Citation of Related Structures:
6CPI, 6CPJ, 6CPK - PubMed Abstract:
Shank proteins are abundant scaffold proteins in the postsynaptic density (PSD) region of brain synapses. Mutations in Shank proteins are associated with autism, schizophrenia, and Alzheimer's disease. To gain insights into Shank protein interactions at the PSD, we determined the solution structures of the src homology 3 (SH3) domains of all three mammalian Shank proteins. Our findings indicate that they have identical and typical SH3 folding motifs, but unusual target-binding pockets. An investigation into the interaction between the Shank SH3 domains and the proline-rich region of the Cav1.3 calcium channel revealed an atypical interaction in which the highly acidic specificity binding pocket of the SH3 domains binds to a Cav1.3 region containing a cluster of three Arg residues. Our study provides insights into Shank SH3-mediated interactions.
Organizational Affiliation:
Department of Biological Sciences, University of Calgary, Canada.