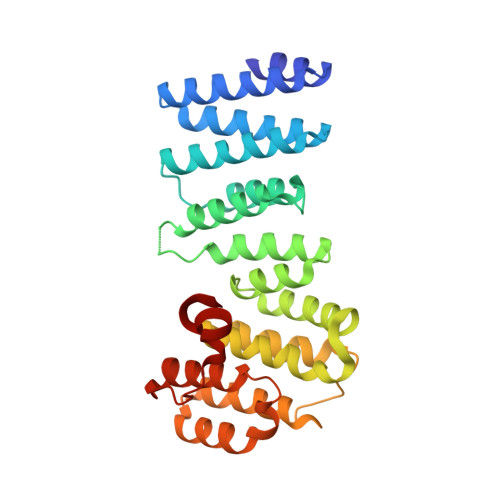
Structural basis for regulation of the nucleo-cytoplasmic distribution of Bag6 by TRC35.
Mock, J.Y., Xu, Y., Ye, Y., Clemons, W.M.(2017) Proc Natl Acad Sci U S A 114: 11679-11684
- PubMed: 29042515
- DOI: https://doi.org/10.1073/pnas.1702940114
- Primary Citation of Related Structures:
6AU8 - PubMed Abstract:
The metazoan protein BCL2-associated athanogene cochaperone 6 (Bag6) forms a hetero-trimeric complex with ubiquitin-like 4A and transmembrane domain recognition complex 35 (TRC35). This Bag6 complex is involved in tail-anchored protein targeting and various protein quality-control pathways in the cytosol as well as regulating transcription and histone methylation in the nucleus. Here we present a crystal structure of Bag6 and its cytoplasmic retention factor TRC35, revealing that TRC35 is remarkably conserved throughout the opisthokont lineage except at the C-terminal Bag6-binding groove, which evolved to accommodate Bag6, a unique metazoan factor. While TRC35 and its fungal homolog, guided entry of tail-anchored protein 4 (Get4), utilize a conserved hydrophobic patch to bind their respective partners, Bag6 wraps around TRC35 on the opposite face relative to the Get4-5 interface. We further demonstrate that TRC35 binding is critical not only for occluding the Bag6 nuclear localization sequence from karyopherin α to retain Bag6 in the cytosol but also for preventing TRC35 from succumbing to RNF126-mediated ubiquitylation and degradation. The results provide a mechanism for regulation of Bag6 nuclear localization and the functional integrity of the Bag6 complex in the cytosol.
Organizational Affiliation:
Division of Chemistry and Chemical Engineering, California Institute of Technology, Pasadena, CA 91125.