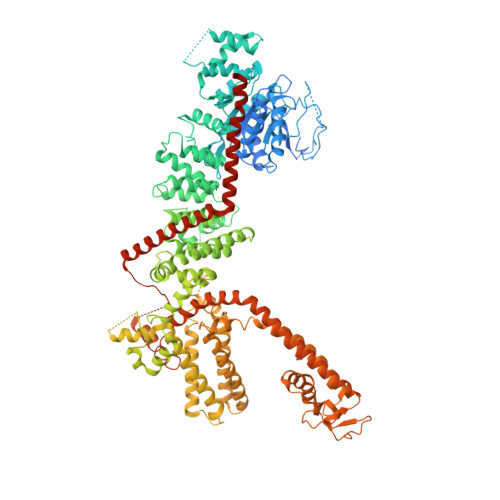
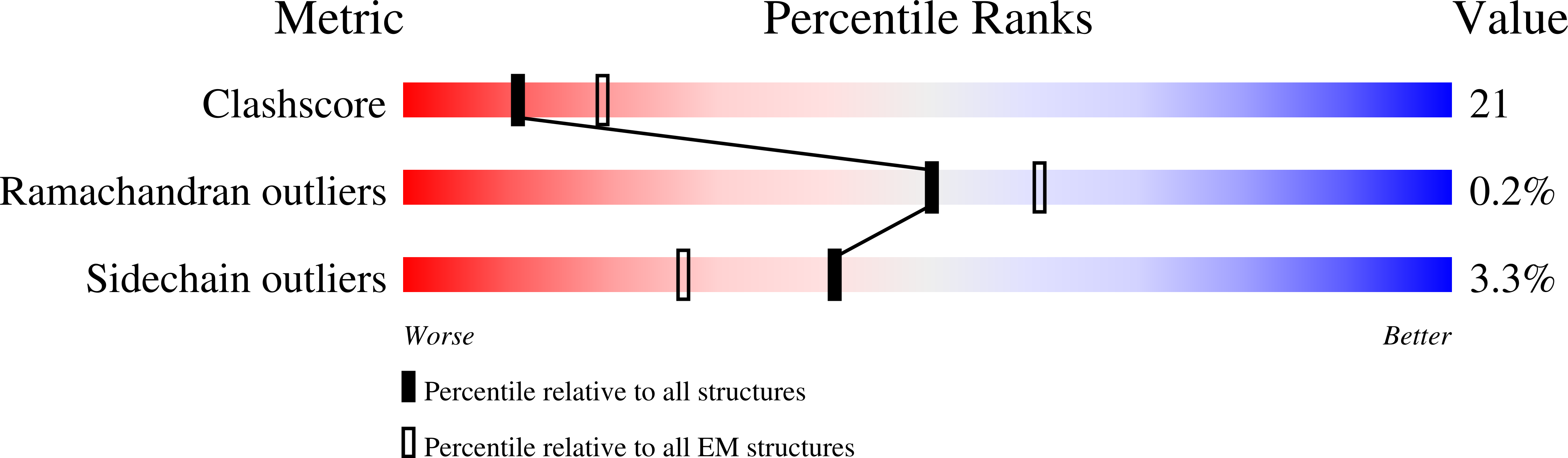Structure of the human TRPM4 ion channel in a lipid nanodisc.
Autzen, H.E., Myasnikov, A.G., Campbell, M.G., Asarnow, D., Julius, D., Cheng, Y.(2018) Science 359: 228-232
- PubMed: 29217581
- DOI: https://doi.org/10.1126/science.aar4510
- Primary Citation of Related Structures:
6BQR, 6BQV - PubMed Abstract:
Transient receptor potential (TRP) melastatin 4 (TRPM4) is a widely expressed cation channel associated with a variety of cardiovascular disorders. TRPM4 is activated by increased intracellular calcium in a voltage-dependent manner but, unlike many other TRP channels, is permeable to monovalent cations only. Here we present two structures of full-length human TRPM4 embedded in lipid nanodiscs at ~3-angstrom resolution, as determined by single-particle cryo-electron microscopy. These structures, with and without calcium bound, reveal a general architecture for this major subfamily of TRP channels and a well-defined calcium-binding site within the intracellular side of the S1-S4 domain. The structures correspond to two distinct closed states. Calcium binding induces conformational changes that likely prime the channel for voltage-dependent opening.
Organizational Affiliation:
Department of Biochemistry and Biophysics, University of California, San Francisco, CA 94143, USA.