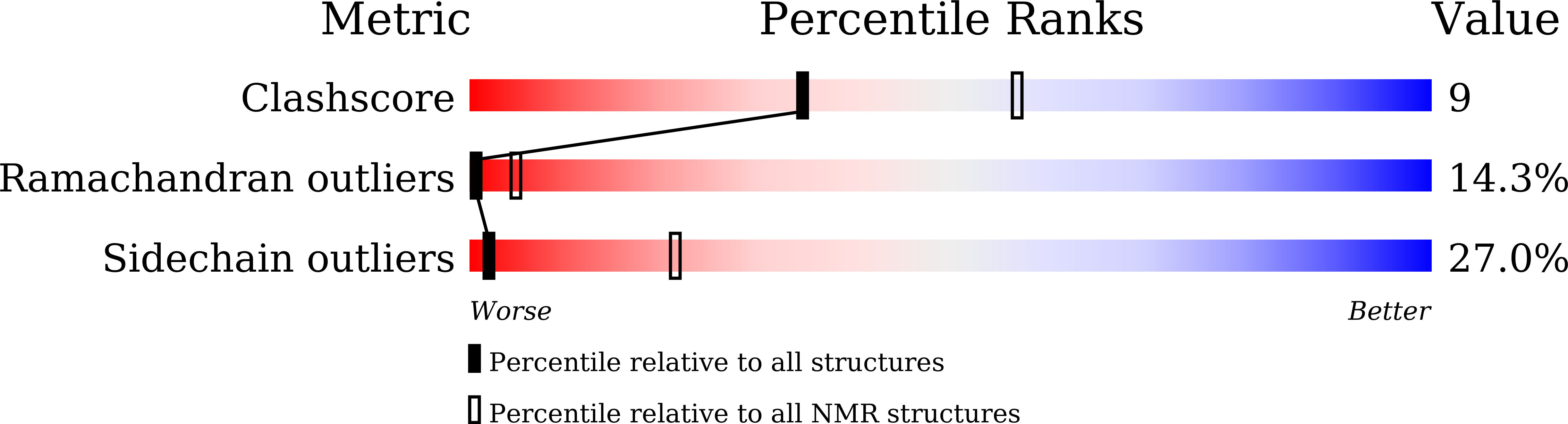Conformational Aspects of High Content Packing of Antimicrobial Peptides in Polymer Microgels
Singh, S., Datta, A., Borro, B.C., Davoudi, M., Schmidtchen, A., Bhunia, A., Malmsten, M.(2017) ACS Appl Mater Interfaces 9: 40094-40106
- PubMed: 29087182
- DOI: https://doi.org/10.1021/acsami.7b13714
- Primary Citation of Related Structures:
5XNG, 5XRX - PubMed Abstract:
Successful use of microgels as delivery systems of antimicrobial peptides (AMPs) requires control of factors determining peptide loading and release to/from the microgels as well as of membrane interactions of both microgel particles and released peptides. Addressing these, we here investigate effects of microgel charge density and conformationally induced peptide amphiphilicity on AMP loading and release using detailed nuclear magnetic resonance (NMR) structural studies combined with ellipsometry, isothermal titration calorimetry, circular dichroism, and light scattering. In parallel, consequences of peptide loading and release for membrane interactions and antimicrobial effects were investigated. In doing so, poly(ethyl acrylate-co-methacrylic acid) microgels were found to incorporate the cationic AMPs EFK17a (EFKRIVQRIKDFLRNLV) and its partially d-amino acid-substituted variant EFK17da (E(dF)KR(dI)VQR(dI)KD(dF)LRNLV). Peptide incorporation was found to increase with increasing with microgel charge density and peptide amphiphilicity. After microgel incorporation, which appeared to occur preferentially in the microgel core, NMR showed EFK17a to form a helix with pronounced amphiphilicity, while EFK17da displayed a folded conformation, stabilized by a hydrophobic hub consisting of aromatic/aromatic and aliphatic/aromatic interactions, resulting in much lower amphiphilicity. Under wide ranges of peptide loading, the microgels displayed net negative z-potential. Such negatively charged microgels do not bind to, nor lyse, bacteria-mimicking membranes. Instead, membrane disruption in these systems is mediated largely by peptide release, which in turn is promoted at higher ionic strength and lower peptide amphiphilicity. Analogously, antimicrobial effects against Escherichia coli were found to be dictated by peptide release. Taken together, the findings show that peptide loading, packing, and release strongly affect the performance of microgels as AMP delivery systems, effects that can be tuned by (conformationally induced) peptide amphiphilicity and by microgel charge density.
Organizational Affiliation:
Department of Pharmacy, Uppsala University , SE-75232 Uppsala, Sweden.