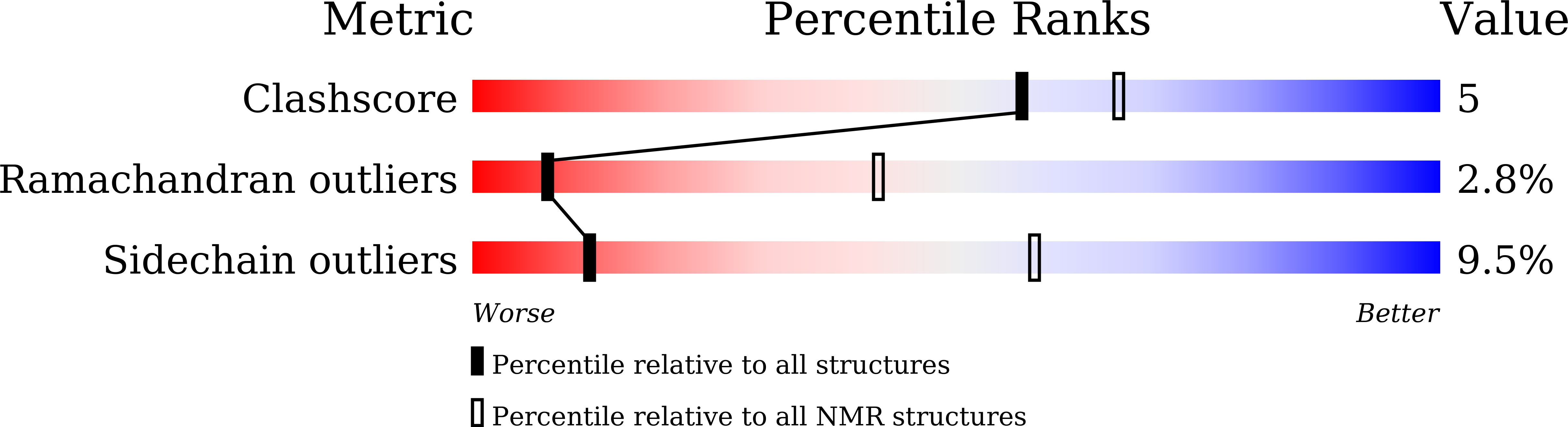Engineering Aromatic-Aromatic Interactions To Nucleate Folding in Intrinsically Disordered Regions of Proteins
Balakrishnan, S., Sarma, S.P.(2017) Biochemistry 56: 4346-4359
- PubMed: 28738155
- DOI: https://doi.org/10.1021/acs.biochem.7b00437
- Primary Citation of Related Structures:
5XE4, 5XEE - PubMed Abstract:
Aromatic interactions are an important force in protein folding as they combine the stability of a hydrophobic interaction with the selectivity of a hydrogen bond. Much of our understanding of aromatic interactions comes from "bioinformatics" based analyses of protein structures and from the contribution of these interactions to stabilizing secondary structure motifs in model peptides. In this study, the structural consequences of aromatic interactions on protein folding have been explored in engineered mutants of the molten globule protein apo-cytochrome b 5 . Structural changes from disorder to order due to aromatic interactions in two variants of the protein, viz., WF-cytb5 and FF-cytb5, result in significant long-range secondary and tertiary structure. The results show that 54 and 52% of the residues in WF-cytb5 and FF-cytb5, respectively, occupy ordered regions versus 26% in apo-cytochrome b 5 . The interactions between the aromatic groups are offset-stacked and edge-to-face for the Trp-Phe and Phe-Phe mutants, respectively. Urea denaturation studies indicate that both mutants have a C m higher than that of apo-cytochrome b 5 and are more stable to chaotropic agents than apo-cytochrome b 5 . The introduction of these aromatic residues also results in "trimer" interactions with existing aromatic groups, reaffirming the selectivity of the aromatic interactions. These studies provide insights into the aromatic interactions that drive disorder-to-order transitions in intrinsically disordered regions of proteins and will aid in de novo protein design beyond small peptide scaffolds.
Organizational Affiliation:
Molecular Biophysics Unit, Indian Institute of Science , Bangalore, Karnataka 560012, India.