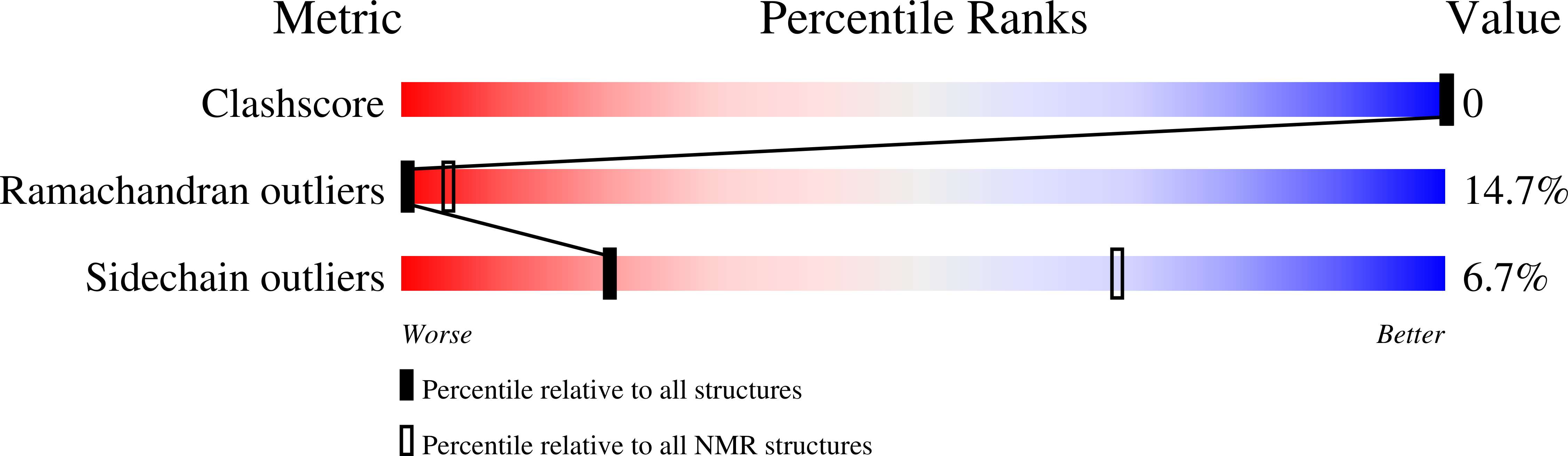Acinetodin and Klebsidin, RNA Polymerase Targeting Lasso Peptides Produced by Human Isolates of Acinetobacter gyllenbergii and Klebsiella pneumoniae.
Metelev, M., Arseniev, A., Bushin, L.B., Kuznedelov, K., Artamonova, T.O., Kondratenko, R., Khodorkovskii, M., Seyedsayamdost, M.R., Severinov, K.(2017) ACS Chem Biol 12: 814-824
- PubMed: 28106375
- DOI: https://doi.org/10.1021/acschembio.6b01154
- Primary Citation of Related Structures:
5UI6, 5UI7 - PubMed Abstract:
We report the bioinformatic prediction and structural validation of two lasso peptides, acinetodin and klebsidin, encoded by the genomes of several human-associated strains of Acinetobacter and Klebsiella. Computation of the three-dimensional structures of these peptides using NMR NOESY constraints verifies that they contain a lasso motif. Despite the lack of sequence similarity to each other or to microcin J25, a prototypical lasso peptide and transcription inhibitor from Escherichia coli, acinetodin and klebsidin also inhibit transcript elongation by the E. coli RNA polymerase by binding to a common site. Yet, unlike microcin J25, acinetodin and klebsidin are unable to permeate wild type E. coli cells and inhibit their growth. We show that the E. coli cells become sensitive to klebsidin when expressing the outer membrane receptor FhuA homologue from Klebsiella pneumoniae. It thus appears that specificity to a common target, the RNA polymerase secondary channel, can be attained by a surprisingly diverse set of primary sequences folded into a common threaded-lasso fold. In contrast, transport into cells containing sensitive targets appears to be much more specific and must be the major determinant of the narrow range of bioactivity of known lasso peptides.
Organizational Affiliation:
Peter the Great St.Petersburg Polytechnic University , St. Petersburg, 195251, Russia.