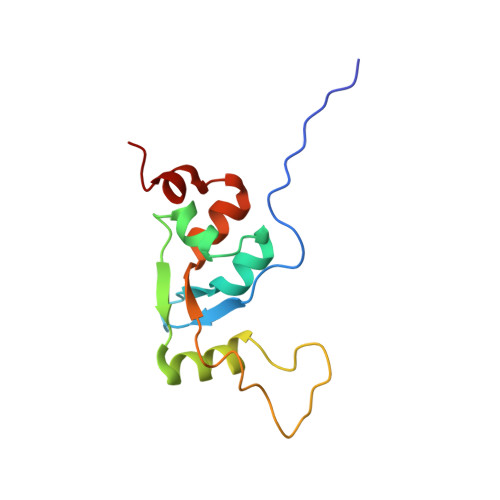
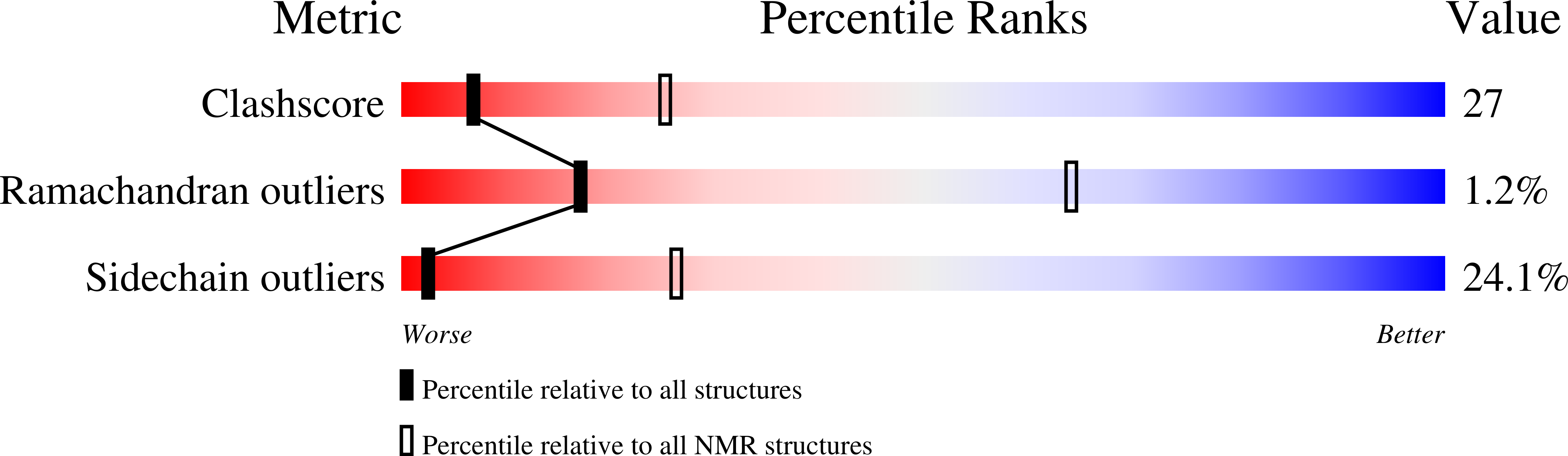The Swi/Snf Subunit Ini1 Contains an N-Terminal Winged Helix DNA Binding Domain that is a Target for Mutations in Schwannomatosis.
Allen, M.D., Freund, S.M.V., Zinzalla, G., Bycroft, M.(2015) Structure 23: 1344
- PubMed: 26073604
- DOI: https://doi.org/10.1016/j.str.2015.04.021
- Primary Citation of Related Structures:
5AJ1 - PubMed Abstract:
SWI/SNF complexes use the energy of ATP hydrolysis to remodel chromatin. In mammals they play a central role in regulating gene expression during differentiation and proliferation. Mutations in SWI/SNF subunits are among the most frequent gene alterations in cancer. The INI1/hSNF5/SMARCB1 subunit is mutated in both malignant rhabdoid tumor, a highly aggressive childhood cancer, and schwannomatosis, a tumor-predisposing syndrome characterized by mostly benign tumors of the CNS. Here, we show that mutations in INI1 that cause schwannomatosis target a hitherto unidentified N-terminal winged helix DNA binding domain that is also present in the BAF45a/PHF10 subunit of the SWI/SNF complex. The domain is structurally related to the SKI/SNO/DAC domain, which is found in a number of metazoan chromatin-associated proteins.
Organizational Affiliation:
MRC Laboratory of Molecular Biology, Francis Crick Avenue, Cambridge Biomedical Campus, Cambridge CB2 0QH, UK.