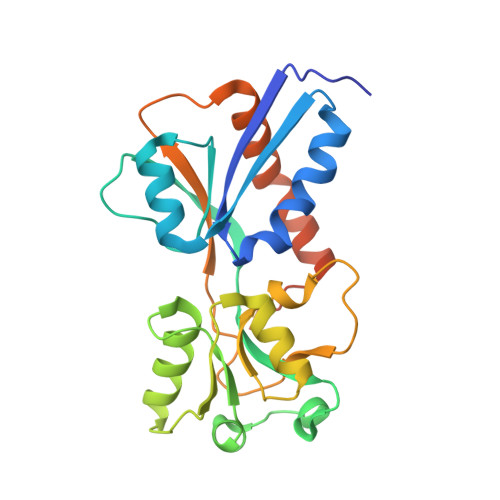
Crystal structure of the effector-binding domain of Synechococcus elongatus CmpR in complex with ribulose 1,5-bisphosphate.
Mahounga, D.M., Sun, H., Jiang, Y.L.(2018) Acta Crystallogr F Struct Biol Commun 74: 506-511
- PubMed: 30084400
- DOI: https://doi.org/10.1107/S2053230X18008841
- Primary Citation of Related Structures:
5Z49 - PubMed Abstract:
The CO 2 -concentrating mechanism (CCM) has evolved to improve the efficiency of photosynthesis in autotrophic cyanobacteria. CmpR, a LysR-type transcriptional regulator (LTTR) from Synechococcus elongatus PCC 7942, was found to regulate CCM-related genes under low-CO 2 conditions. Here, the dimeric structure of the effector-binding domain of CmpR (CmpR-EBD) in complex with the co-activator ribulose 1,5-bisphosphate (RuBP) is reported at 2.15 Å resolution. One RuBP molecule binds to the inter-domain cleft between the two subunits of the CmpR-EBD dimer. Structural comparison combined with sequence analyses demonstrated that CmpR-EBD has an overall structure similar to those of LTTRs of known structure, but possesses a distinctly different effector-binding pattern.
Organizational Affiliation:
School of Life Sciences and Hefei National Laboratory for Physical Sciences at the Microscale, University of Science and Technology of China, Hefei, Anhui 230027, People's Republic of China.