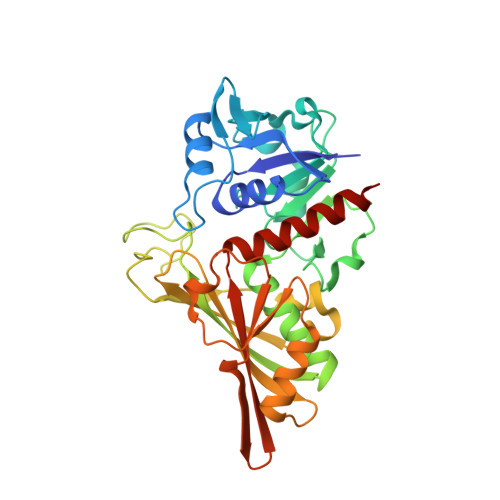
High-resolution crystal structure of Streptococcus agalactiae glyceraldehyde-3-phosphate dehydrogenase.
Zhou, K., Fan, X., Li, Y., Zhang, C., Jin, T.(2018) Acta Crystallogr F Struct Biol Commun 74: 236-244
- PubMed: 29633972
- DOI: https://doi.org/10.1107/S2053230X18003801
- Primary Citation of Related Structures:
5Y37 - PubMed Abstract:
Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) is a multifunctional enzyme that plays critical roles in bacterial pathogenesis in some pathogenic bacteria. In this study, the crystal structure of group B streptococcus GAPDH was determined at 1.36 Å resolution. The structure contained an asymmetric mixed holo tetramer, with two NAD ligands bound to two protomers. Further structural analysis identified interesting phosphate ion-binding sites, which shed light on its catalytic mechanism.
Organizational Affiliation:
Laboratory of Structural Immunology, CAS Key Laboratory of Innate Immunity and Chronic Disease, CAS Center for Excellence in Molecular Cell Science, School of Life Sciences and Medical Center, University of Science and Technology of China, Hefei, Anhui 230027, People's Republic of China.