Potent and selective inhibition of pathogenic viruses by engineered ubiquitin variants.
Zhang, W., Bailey-Elkin, B.A., Knaap, R.C.M., Khare, B., Dalebout, T.J., Johnson, G.G., van Kasteren, P.B., McLeish, N.J., Gu, J., He, W., Kikkert, M., Mark, B.L., Sidhu, S.S.(2017) PLoS Pathog 13: e1006372-e1006372
- PubMed: 28542609
- DOI: https://doi.org/10.1371/journal.ppat.1006372
- Primary Citation of Related Structures:
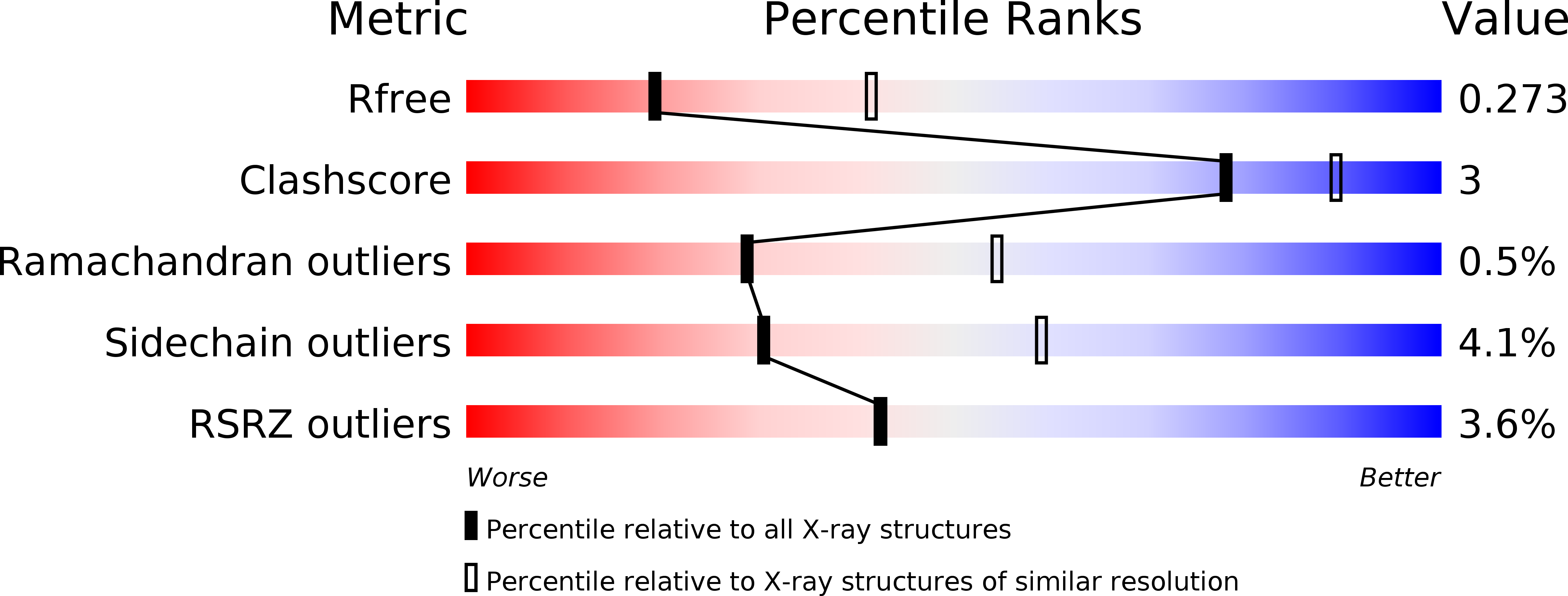
5V5G, 5V5H, 5V69, 5V6A - PubMed Abstract:
The recent Middle East respiratory syndrome coronavirus (MERS-CoV), Ebola and Zika virus outbreaks exemplify the continued threat of (re-)emerging viruses to human health, and our inability to rapidly develop effective therapeutic countermeasures. Many viruses, including MERS-CoV and the Crimean-Congo hemorrhagic fever virus (CCHFV) encode deubiquitinating (DUB) enzymes that are critical for viral replication and pathogenicity. They bind and remove ubiquitin (Ub) and interferon stimulated gene 15 (ISG15) from cellular proteins to suppress host antiviral innate immune responses. A variety of viral DUBs (vDUBs), including the MERS-CoV papain-like protease, are responsible for cleaving the viral replicase polyproteins during replication, and are thereby critical components of the viral replication cycle. Together, this makes vDUBs highly attractive antiviral drug targets. However, structural similarity between the catalytic cores of vDUBs and human DUBs complicates the development of selective small molecule vDUB inhibitors. We have thus developed an alternative strategy to target the vDUB activity through a rational protein design approach. Here, we report the use of phage-displayed ubiquitin variant (UbV) libraries to rapidly identify potent and highly selective protein-based inhibitors targeting the DUB domains of MERS-CoV and CCHFV. UbVs bound the vDUBs with high affinity and specificity to inhibit deubiquitination, deISGylation and in the case of MERS-CoV also viral replicative polyprotein processing. Co-crystallization studies further revealed critical molecular interactions between UbVs and MERS-CoV or CCHFV vDUBs, accounting for the observed binding specificity and high affinity. Finally, expression of UbVs during MERS-CoV infection reduced infectious progeny titers by more than four orders of magnitude, demonstrating the remarkable potency of UbVs as antiviral agents. Our results thereby establish a strategy to produce protein-based inhibitors that could protect against a diverse range of viruses by providing UbVs via mRNA or protein delivery technologies or through transgenic techniques.
Organizational Affiliation:
Donnelly Centre for Cellular and Biomolecular Research, Banting and Best Department of Medical Research, and Department of Molecular Genetics, University of Toronto, Toronto, Ontario, Canada.