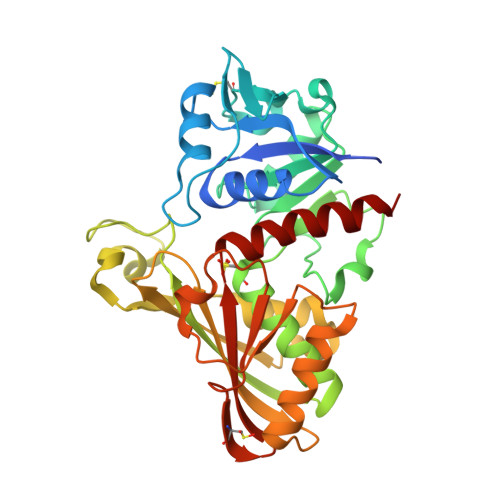
Structural studies of glyceraldehyde-3-phosphate dehydrogenase from Naegleria gruberi, the first one from phylum Percolozoa.
Machado, A.T.P., Silva, M., Iulek, J.(2018) Biochim Biophys Acta 1866: 581-588
- PubMed: 29501559
- DOI: https://doi.org/10.1016/j.bbapap.2018.02.006
- Primary Citation of Related Structures:
5UR0 - PubMed Abstract:
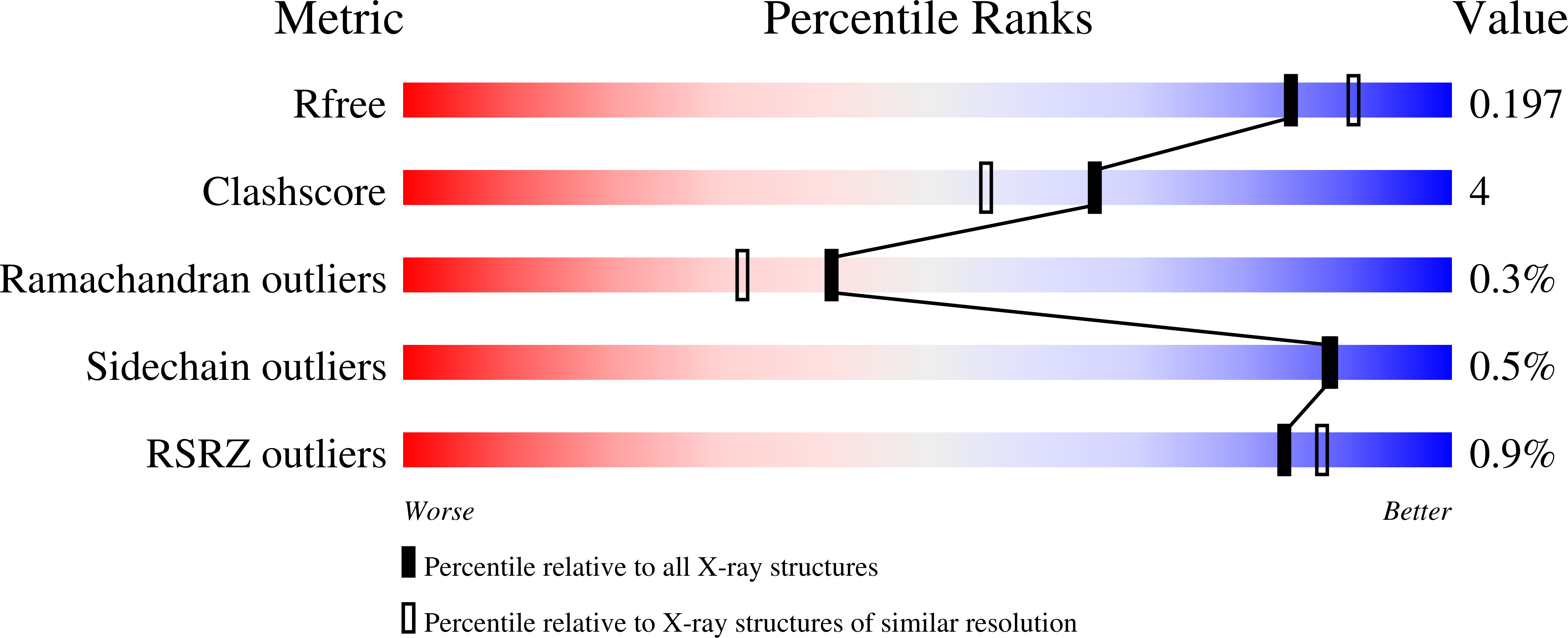
Naegleria gruberi is a free life amoeba believed to have more than one billion years of existence; it is not pathogenic and had its genome sequenced, which revealed a high complexity in the metabolic pathways. This paper presents the experimental structure of GAPDH from N. gruberi, the first one belonging to the phylum Percolozoa, comparisons to structures from various species and molecular dynamics studies of some particular features. The final refined structure presents R cryst = 15.54% and R free = 19.84%. The catalytic domain formed by residues 134 to 313 is highly conserved, as expected, with the exception of Asn145, present only in NgGAPDH, while the other GAPDHs present either Ser or Thr on the corresponding position. Molecular dynamics analysis revealed that Asn145 has correlated motions with residues Ala123, Thr125 and Pro126 that belong to what was called "bonded loop". NgGAPDH residue Met35 presents an extended side chain, closer to the cofactor adenine ring than corresponding (different) residues and conformations found in some parasitic protozoa and the human GAPDHs. The enzyme was previously reported to present positive cooperativity, which is hypothesized to be related to certain atom distances.
Organizational Affiliation:
Chemistry, State University of Ponta Grossa, Ponta Grossa 84030-900, Brazil.