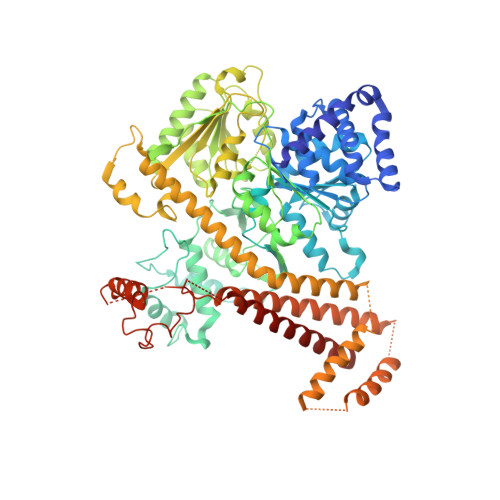
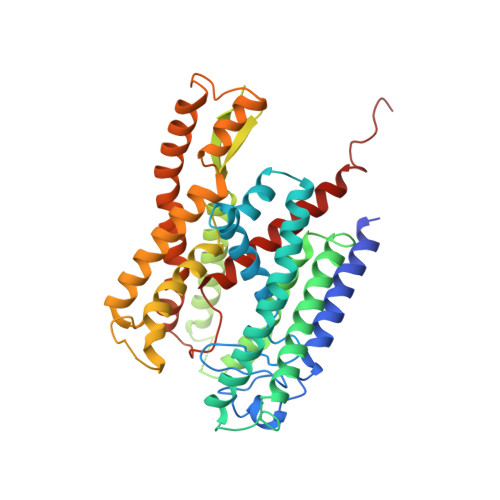
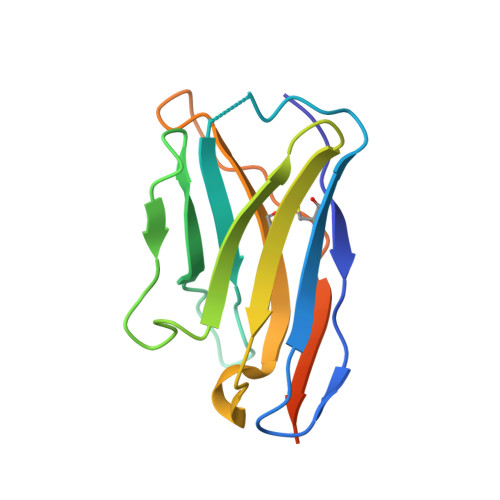
Crystal structure of a substrate-engaged SecY protein-translocation channel.
Li, L., Park, E., Ling, J., Ingram, J., Ploegh, H., Rapoport, T.A.(2016) Nature 531: 395-399
- PubMed: 26950603
- DOI: https://doi.org/10.1038/nature17163
- Primary Citation of Related Structures:
5EUL - PubMed Abstract:
Hydrophobic signal sequences target secretory polypeptides to a protein-conducting channel formed by a heterotrimeric membrane protein complex, the prokaryotic SecY or eukaryotic Sec61 complex. How signal sequences are recognized is poorly understood, particularly because they are diverse in sequence and length. Structures of the inactive channel show that the largest subunit, SecY or Sec61α, consists of two halves that form an hourglass-shaped pore with a constriction in the middle of the membrane and a lateral gate that faces lipid. The cytoplasmic funnel is empty, while the extracellular funnel is filled with a plug domain. In bacteria, the SecY channel associates with the translating ribosome in co-translational translocation, and with the SecA ATPase in post-translational translocation. How a translocating polypeptide inserts into the channel is uncertain, as cryo-electron microscopy structures of the active channel have a relatively low resolution (~10 Å) or are of insufficient quality. Here we report a crystal structure of the active channel, assembled from SecY complex, the SecA ATPase, and a segment of a secretory protein fused into SecA. The translocating protein segment inserts into the channel as a loop, displacing the plug domain. The hydrophobic core of the signal sequence forms a helix that sits in a groove outside the lateral gate, while the following polypeptide segment intercalates into the gate. The carboxy (C)-terminal section of the polypeptide loop is located in the channel, surrounded by residues of the pore ring. Thus, during translocation, the hydrophobic segments of signal sequences, and probably bilayer-spanning domains of nascent membrane proteins, exit the lateral gate and dock at a specific site that faces the lipid phase.
- Howard Hughes Medical Institute and Harvard Medical School, Department of Cell Biology, 240 Longwood Avenue, Boston, MA 02115, USA.
Organizational Affiliation: