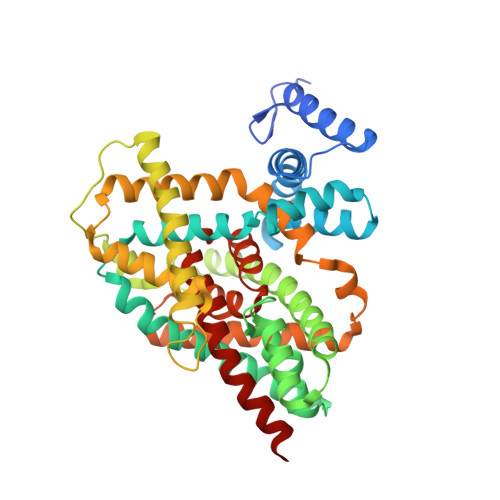
Mechanism of Na(+)-dependent citrate transport from the structure of an asymmetrical CitS dimer.
Wohlert, D., Grotzinger, M.J., Kuhlbrandt, W., Yildiz, O.(2015) Elife 4: e09375-e09375
- PubMed: 26636752
- DOI: https://doi.org/10.7554/eLife.09375
- Primary Citation of Related Structures:
5A1S - PubMed Abstract:
The common human pathogen Salmonella enterica takes up citrate as a nutrient via the sodium symporter SeCitS. Uniquely, our 2.5 Å x-ray structure of the SeCitS dimer shows three different conformations of the active protomer. One protomer is in the outside-facing state. Two are in different inside-facing states. All three states resolve the substrates in their respective binding environments. Together with comprehensive functional studies on reconstituted proteoliposomes, the structures explain the transport mechanism in detail. Our results indicate a six-step process, with a rigid-body 31° rotation of a helix bundle that translocates the bound substrates by 16 Å across the membrane. Similar transport mechanisms may apply to a wide variety of related and unrelated secondary transporters, including important drug targets.
- Department of Structural Biology, Max Planck Institute of Biophysics, Frankfurt am Main, Germany.
Organizational Affiliation: