Naphthalene-sulfonate inhibitors of human norovirus RNA-dependent RNA-polymerase.
Tarantino, D., Pezzullo, M., Mastrangelo, E., Croci, R., Rohayem, J., Robel, I., Bolognesi, M., Milani, M.(2014) Antiviral Res 102: 23-28
- PubMed: 24316032
- DOI: https://doi.org/10.1016/j.antiviral.2013.11.016
- Primary Citation of Related Structures:
4LQ3, 4LQ9 - PubMed Abstract:
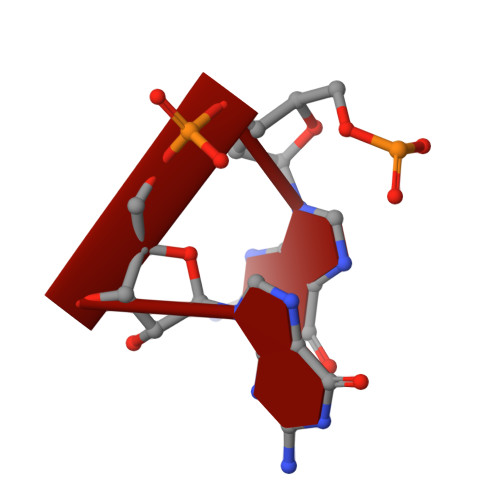
Noroviruses are members of the Caliciviridae family of positive sense RNA viruses. In humans Noroviruses cause rapid onset diarrhea and vomiting. Currently Norovirus infection is responsible for 21 million gastroenteritis yearly cases in the USA. Nevertheless, despite the obvious public health and socio-economic relevance, no effective vaccines/antivirals are yet available to treat Norovirus infection. Since the activity of RNA-dependent RNA polymerase (RdRp) plays a key role in genome replication and in the synthesis/amplification of subgenomic RNA, the enzyme is considered a promising target for antiviral drug development. In this context, following the identification of suramin and NF023 as Norovirus RdRp inhibitors, we analyzed the potential inhibitory role of naphthalene di-sulfonate (NAF2), a fragment derived from these two molecules. Although NAF2, tested in enzymatic polymerase inhibition assays, displayed low activity against RdRp (IC50=14μM), the crystal structure of human Norovirus RdRp revealed a thumb domain NAF2 binding site that differs from that characterized for NF023/suramin. To further map the new potential inhibitory site, we focused on the structurally related molecule pyridoxal-5'-phosphate-6-(2'-naphthylazo-6'-nitro-4',8'-disulfonate) tetrasodium salt (PPNDS). PPNDS displayed below-micromolar inhibitory activity versus human Norovirus RdRp (IC50=0.45μM), similarly to suramin and NF023. Inspection of the crystal structure of the RdRp/PPNDS complex showed that the inhibitor bound to the NAF2 thumb domain site, highlighting the relevance of such new binding site for exploiting Norovirus RdRp inhibitors.
Organizational Affiliation:
Department of Biosciences and CIMAINA, University of Milano, Via Celoria 26, I-20133 Milano, Italy.