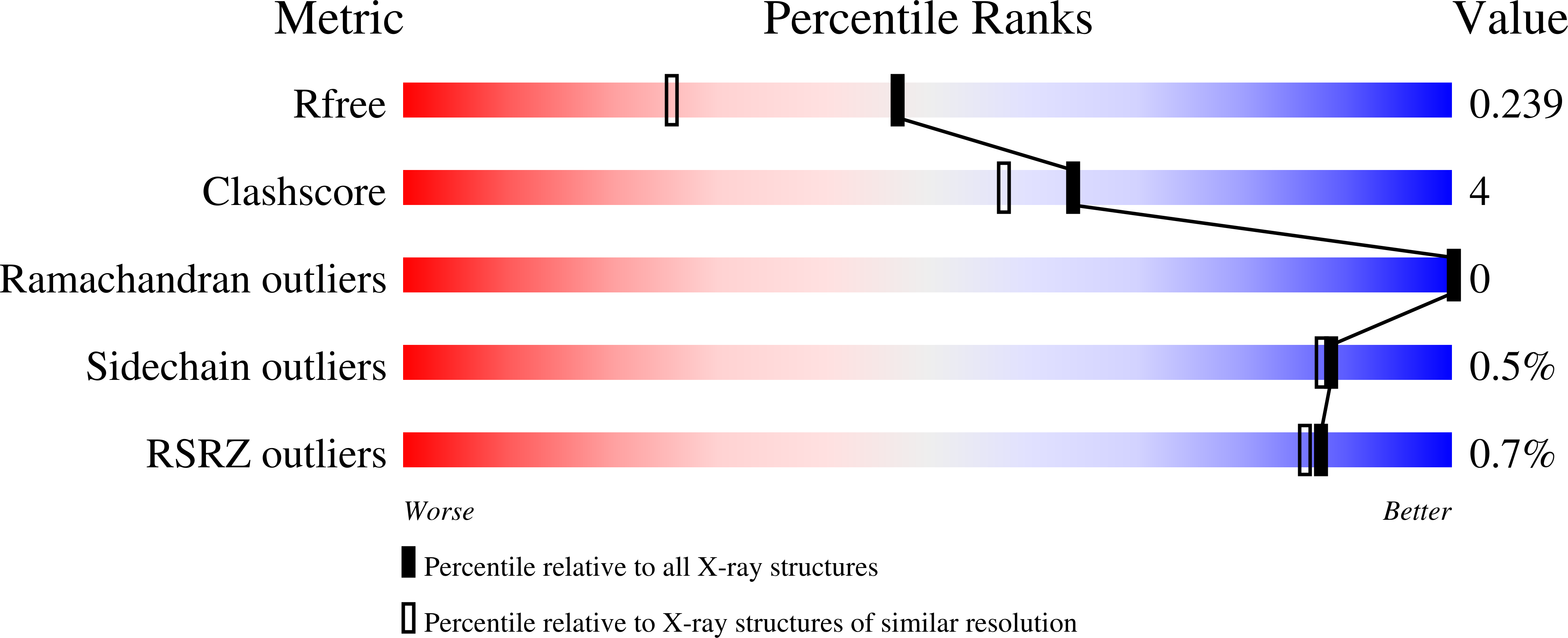
Complexes of dual-function hemoglobin/dehaloperoxidase with substrate 2,4,6-trichlorophenol are inhibitory and indicate binding of halophenol to compound I.
Wang, C., Lovelace, L.L., Sun, S., Dawson, J.H., Lebioda, L.(2013) Biochemistry 52: 6203-6210
- PubMed: 23952341
- DOI: https://doi.org/10.1021/bi400627w
- Primary Citation of Related Structures:
4KMV, 4KMW, 4KN3 - PubMed Abstract:
The hemoglobin of sea worm Amphitrite ornata, which for historical reasons is abbreviated as DHP for dehaloperoxidase, has two physiological functions: it binds dioxygen in the ferrous state and dehalogenates halophenols, such as 2,4,6-trichlorophenol (TCP), using hydrogen peroxide as the oxidant in the ferric state. The crystal structures of three DHP variants (Y34N, Y34N/S91G, and L100F) with TCP bound show two mutually exclusive modes of substrate binding. One of them, the internal site, is deep inside the distal pocket with the phenolic OH moiety forming a hydrogen bond to the water molecule coordinated to the heme Fe. In this complex, the distal histidine is predominantly located in the closed position and also forms a hydrogen bond to the phenolic hydroxide. The second mode of TCP binding is external, at the heme edge, with the halophenol molecule forming a lid covering the entrance to the distal cavity. The distal histidine is in the open position and forms a hydrogen bond to the OH group of TCP, which also hydrogen bonds to the hydroxyl of Tyr38. The distance between the Cl4 atom of TCP and the heme Fe is 3.9 Å (nonbonding). In both complexes, TCP molecules prevent the approach of hydrogen peroxide to the heme, indicating that the complexes are inhibitory and implying that the substrates must bind in an ordered fashion: hydrogen peroxide first and TCP second. Kinetic studies confirmed the inhibition of DHP by high concentrations of TCP. The external binding mode may resemble the interaction of TCP with Compound I, the catalytic intermediate to which halophenols bind. The measured values of the apparent Km for TCP were in the range of 0.3-0.8 mM, much lower than the concentrations required to observe TCP binding in crystals. This indicates that during catalysis TCP binds to Compound I. Mutant F21W, which likely has the internal TCP binding site blocked, has ~7% of the activity of wild-type DHP.
Organizational Affiliation:
Department of Chemistry and Biochemistry, University of South Carolina , Columbia, South Carolina 29208, United States.