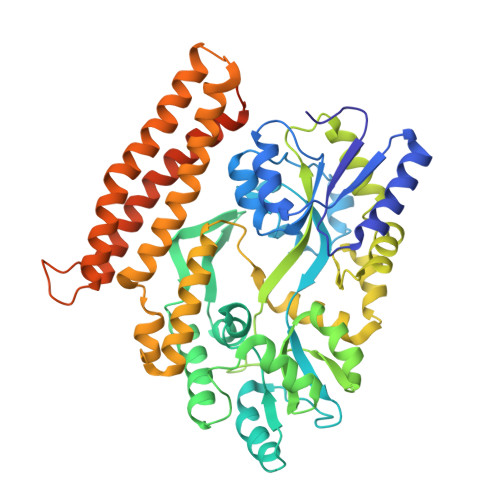
Structure of the GH1 domain of guanylate kinase-associated protein from Rattus norvegicus.
Tong, J., Yang, H., Eom, S.H., Chun, C., Im, Y.J.(2014) Biochem Biophys Res Commun 452: 130-135
- PubMed: 25152391
- DOI: https://doi.org/10.1016/j.bbrc.2014.08.073
- Primary Citation of Related Structures:
4R0Y - PubMed Abstract:
Guanylate-kinase-associated protein (GKAP) is a scaffolding protein that links NMDA receptor-PSD-95 to Shank-Homer complexes by protein-protein interactions at the synaptic junction. GKAP family proteins are characterized by the presence of a C-terminal conserved GKAP homology domain 1 (GH1) of unknown structure and function. In this study, crystal structure of the GH1 domain of GKAP from Rattus norvegicus was determined in fusion with an N-terminal maltose-binding protein at 2.0 Å resolution. The structure of GKAP GH1 displays a three-helix bundle connected by short flexible loops. The predicted helix α4 which was not visible in the crystal structure associates weakly with the helix α3 suggesting dynamic nature of the GH1 domain. The strict conservation of GH1 domain across GKAP family members and the lack of a catalytic active site required for enzyme activity imply that the GH1 domain might serve as a protein-protein interaction module for the synaptic protein clustering.
Organizational Affiliation:
College of Pharmacy, Chonnam National University, Gwangju 500-757, South Korea.