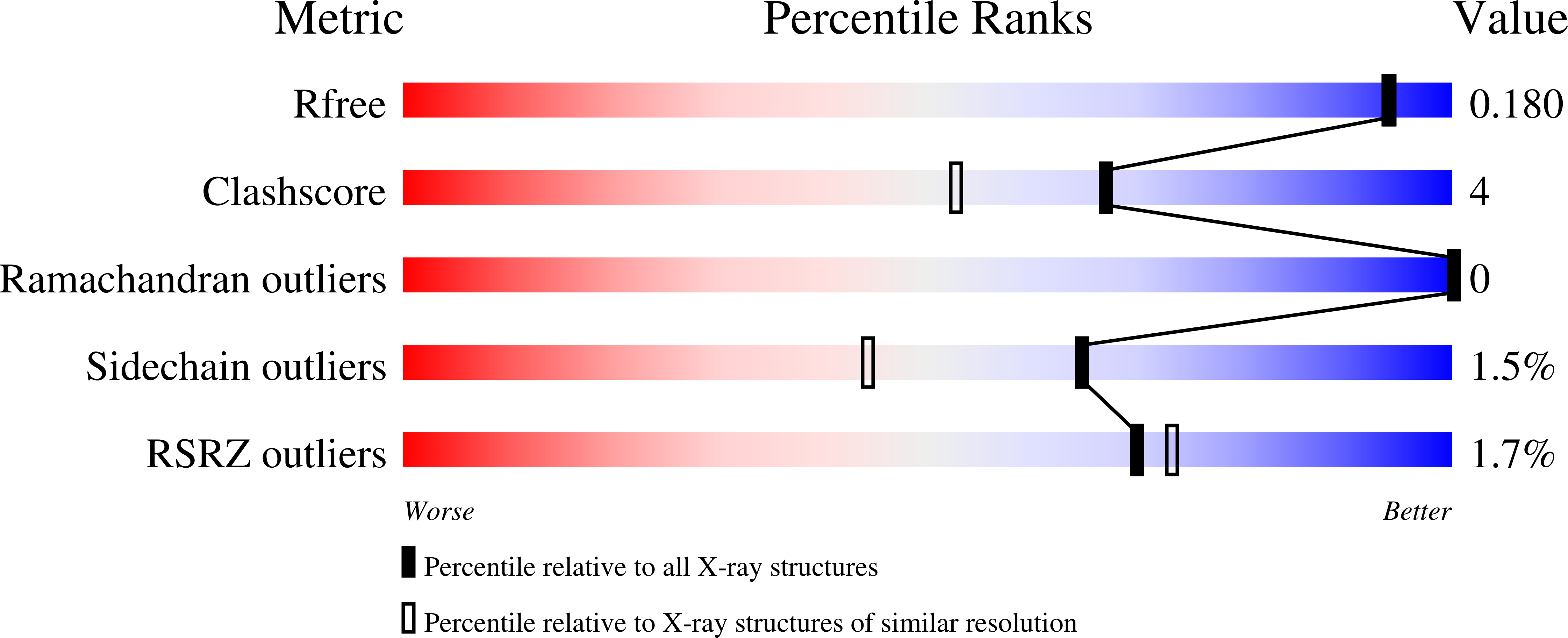
Crystal structure of the phosphate-binding protein (PBP-1) of an ABC-type phosphate transporter from Clostridium perfringens.
Gonzalez, D., Richez, M., Bergonzi, C., Chabriere, E., Elias, M.(2014) Sci Rep 4: 6636-6636
- PubMed: 25338617
- DOI: https://doi.org/10.1038/srep06636
- Primary Citation of Related Structures:
4Q8R - PubMed Abstract:
Phosphate limitation is an important environmental stress that affects the metabolism of various organisms and, in particular, can trigger the virulence of numerous bacterial pathogens. Clostridium perfringens, a human pathogen, is one of the most common causes of enteritis necroticans, gas gangrene and food poisoning. Here, we focused on the high affinity phosphate-binding protein (PBP-1) of an ABC-type transporter, responsible for cellular phosphate uptake. We report the crystal structure (1.65 Å resolution) of the protein in complex with phosphate. Interestingly, PBP-1 does not form the short, low-barrier hydrogen bond with phosphate that is typical of previously characterized phosphate-binding proteins, but rather a canonical hydrogen bond. In its unique binding configuration, PBP-1 forms an unusually high number of hydrogen bonds (14) with the phosphate anion. Discrimination experiments reveal that PBP-1 is the least selective PBP characterised so far and is able to discriminate phosphate from its close competing anion, arsenate, by ~150-fold.
Organizational Affiliation:
URMITE UMR CNRS-IRD 6236, IFR48, Faculté de Médecine et de Pharmacie, Université de la Méditerranée, Marseille, France.