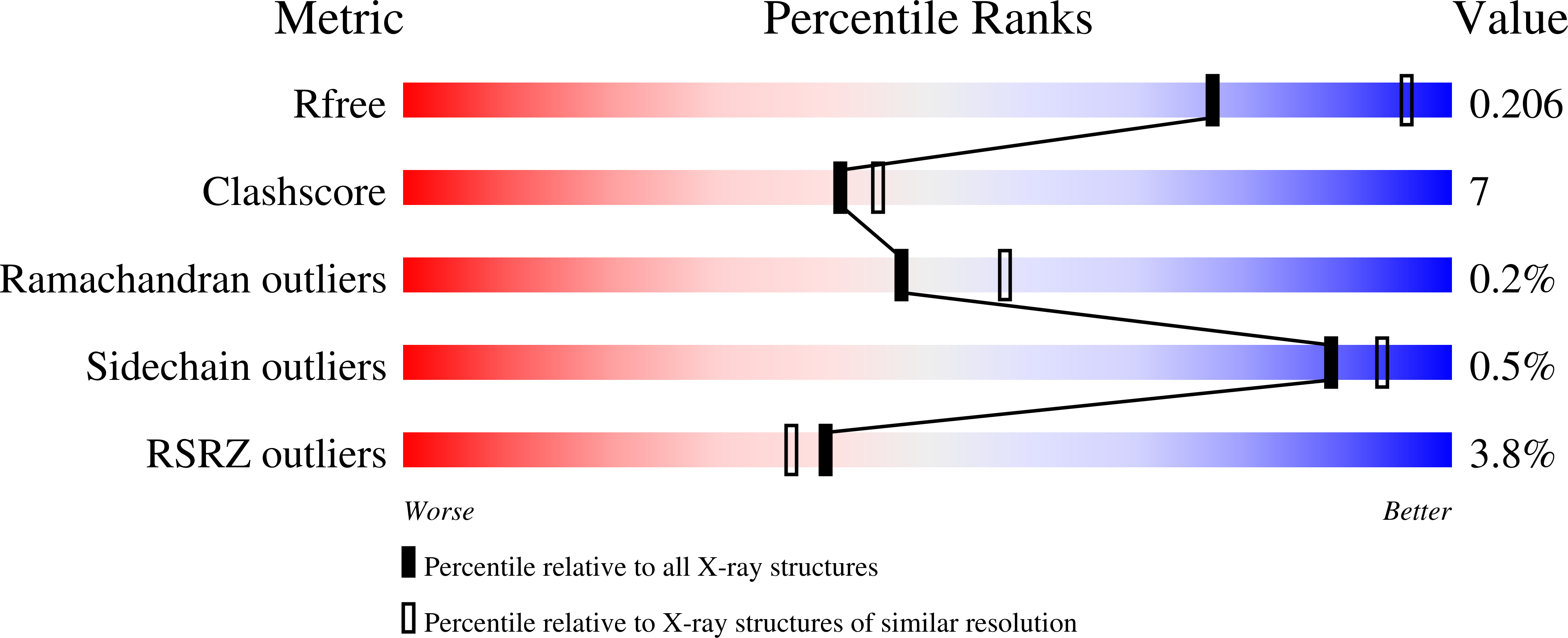
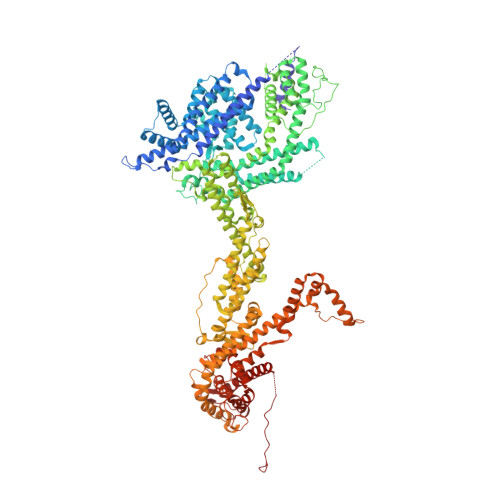
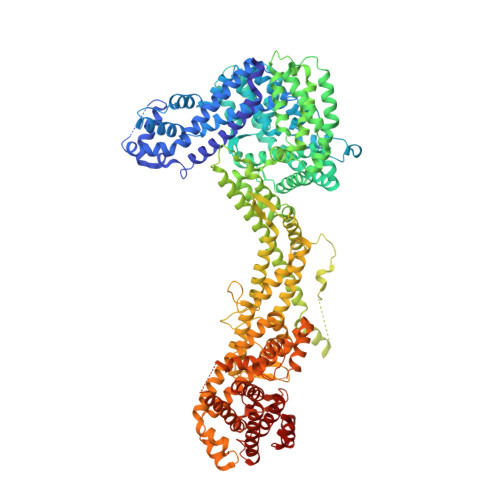
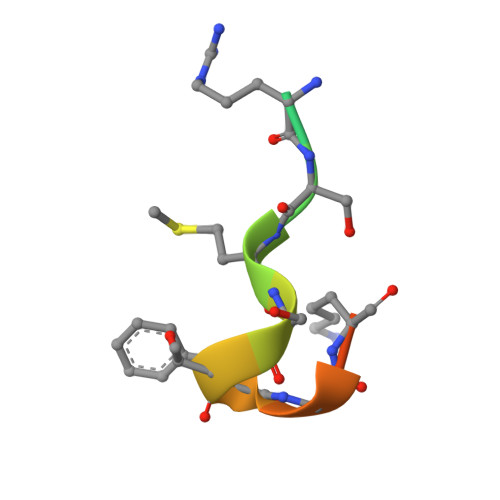
The WAVE Regulatory Complex Links Diverse Receptors to the Actin Cytoskeleton.
Chen, B., Brinkmann, K., Chen, Z., Pak, C.W., Liao, Y., Shi, S., Henry, L., Grishin, N.V., Bogdan, S., Rosen, M.K.(2014) Cell 156: 195-207
- PubMed: 24439376
- DOI: https://doi.org/10.1016/j.cell.2013.11.048
- Primary Citation of Related Structures:
4N78 - PubMed Abstract:
The WAVE regulatory complex (WRC) controls actin cytoskeletal dynamics throughout the cell by stimulating the actin-nucleating activity of the Arp2/3 complex at distinct membrane sites. However, the factors that recruit the WRC to specific locations remain poorly understood. Here, we have identified a large family of potential WRC ligands, consisting of ∼120 diverse membrane proteins, including protocadherins, ROBOs, netrin receptors, neuroligins, GPCRs, and channels. Structural, biochemical, and cellular studies reveal that a sequence motif that defines these ligands binds to a highly conserved interaction surface of the WRC formed by the Sra and Abi subunits. Mutating this binding surface in flies resulted in defects in actin cytoskeletal organization and egg morphology during oogenesis, leading to female sterility. Our findings directly link diverse membrane proteins to the WRC and actin cytoskeleton and have broad physiological and pathological ramifications in metazoans.
Organizational Affiliation:
Department of Biophysics and Howard Hughes Medical Institute, University of Texas Southwestern Medical Center at Dallas, 5323 Harry Hines Boulevard, Dallas, TX 75390, USA.