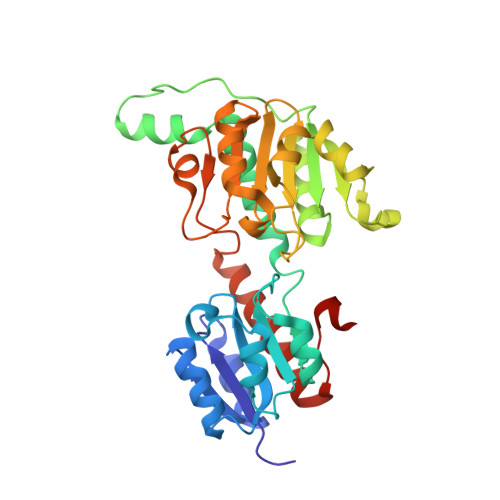
Characterization and crystal structure of a first fungal glyoxylate reductase from Paecilomyes thermophila
Duan, X., Hu, S., Zhou, P., Zhou, Y., Liu, Y., Jiang, Z.(2014) Enzyme Microb Technol 60: 72-79
- PubMed: 24835102
- DOI: https://doi.org/10.1016/j.enzmictec.2014.04.004
- Primary Citation of Related Structures:
3WNV - PubMed Abstract:
A glyoxylate reductase gene (PtGR) from the fungus Paecilomyces thermophila was cloned and expressed in Escherichia coli. PtGR was biochemically and structurally characterized. PtGR has an open reading frame of 993bp encoding 330 amino acids. The deduced amino acid sequence has low similarities to the reported glyoxylate reductases. The purified PtGR forms a homodimer. PtGR displayed an optimum pH of 7.5 and broad pH stability (pH 4.5-10). It exhibited an optimal temperature of 50°C and was stable up to 50°C. PtGR was found to be highly specific for glyoxylate, but it showed no detectable activity with 4-methyl-2-oxopentanoate, phenylglyoxylate, pyruvate, oxaloacetate and α-ketoglutarate. PtGR prefered NADPH rather than NADH as an electron donor. Moreover, the crystal structure of PtGR was determined at 1.75Å resolution. The overall structure of apo-PtGR monomer adopts the typical d-2-hydroxy-acid dehydrogenase fold with a "closed" conformation unexpectedly. The coenzyme specificity is provided by a cationic cluster consisting of N184, R185, and N186 structurally. These structural observations could explain its different coenzyme and substrate specificity.
Organizational Affiliation:
Department of Biotechnology, College of Food Science and Nutritional Engineering, China Agricultural University, Beijing 100083, China.