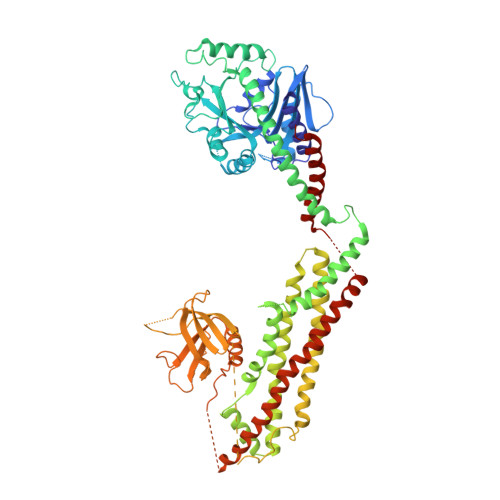
Crystal structure of nucleotide-free dynamin.
Faelber, K., Posor, Y., Gao, S., Held, M., Roske, Y., Schulze, D., Haucke, V., Noe, F., Daumke, O.(2011) Nature 477: 556-560
- PubMed: 21927000
- DOI: https://doi.org/10.1038/nature10369
- Primary Citation of Related Structures:
3SNH - PubMed Abstract:
Dynamin is a mechanochemical GTPase that oligomerizes around the neck of clathrin-coated pits and catalyses vesicle scission in a GTP-hydrolysis-dependent manner. The molecular details of oligomerization and the mechanism of the mechanochemical coupling are currently unknown. Here we present the crystal structure of human dynamin 1 in the nucleotide-free state with a four-domain architecture comprising the GTPase domain, the bundle signalling element, the stalk and the pleckstrin homology domain. Dynamin 1 oligomerized in the crystals via the stalks, which assemble in a criss-cross fashion. The stalks further interact via conserved surfaces with the pleckstrin homology domain and the bundle signalling element of the neighbouring dynamin molecule. This intricate domain interaction rationalizes a number of disease-related mutations in dynamin 2 and suggests a structural model for the mechanochemical coupling that reconciles previous models of dynamin function.
Organizational Affiliation:
Crystallography, Max-Delbrück-Centrum for Molecular Medicine, Robert-Rössle-Strasse 10, 13125 Berlin, Germany. katja.faelber@mdc-berlin.de