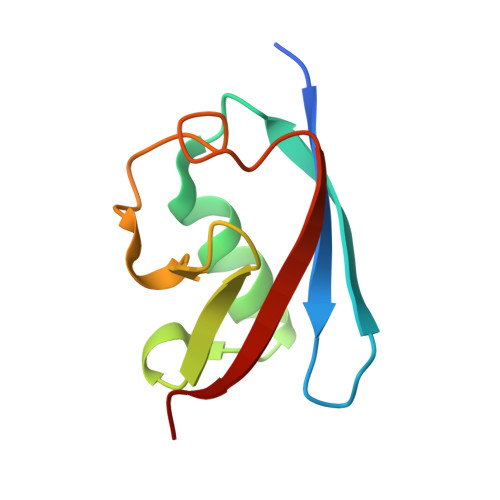
Crystal structure of human FAF1 UBX domain reveals a novel FcisP touch-turn motif in p97/VCP-binding region
Kang, W., Yang, J.K.(2011) Biochem Biophys Res Commun 407: 531-534
- PubMed: 21414298
- DOI: https://doi.org/10.1016/j.bbrc.2011.03.052
- Primary Citation of Related Structures:
3QCA - PubMed Abstract:
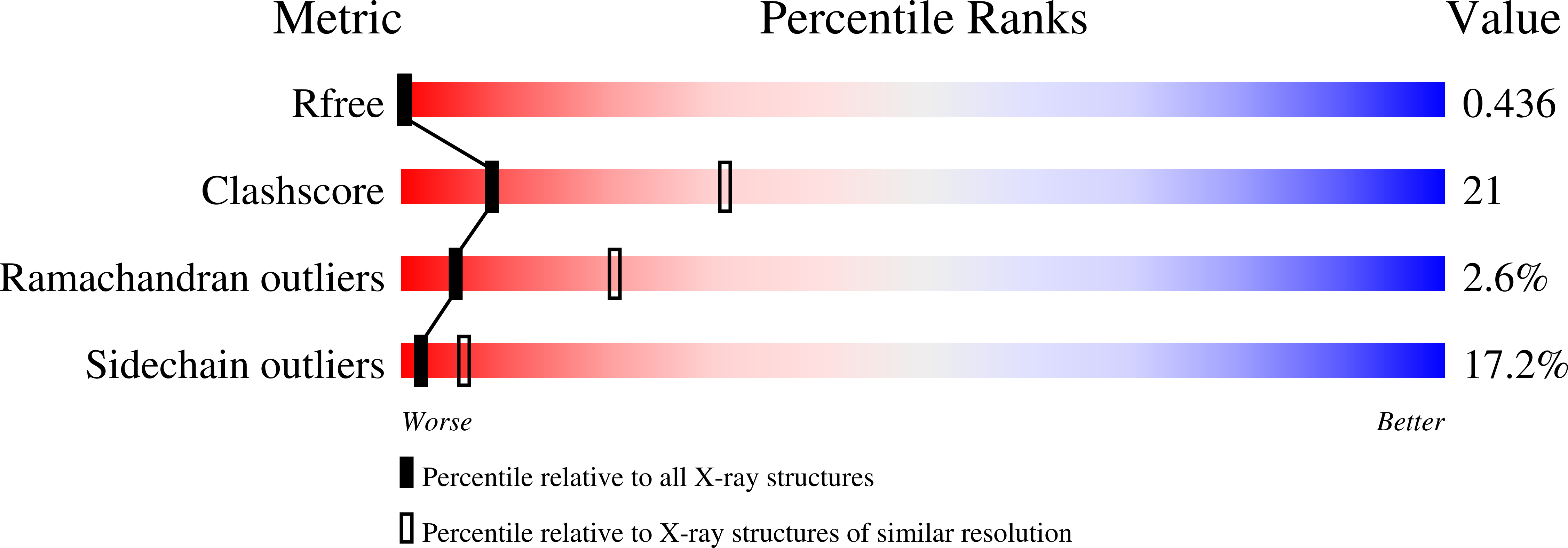
UBX domain is a general p97/VCP-binding module found in an increasing number of proteins including FAF1, p47, SAKS1 and UBXD7. FAF1, a multi-functional tumor suppressor protein, binds to the N domain of p97/VCP through its C-terminal UBX domain and thereby inhibits the proteasomal protein degradation in which p97/VCP acts as a co-chaperone. Here we report the crystal structure of human FAF1 UBX domain at 2.9Å resolution. It reveals that the conserved FP sequence in the p97/VCP-binding region adopts a rarely observed cis-Pro touch-turn structure. We call it an FcisP touch-turn motif and suggest that it is the conserved structural element of the UBX domain. Four FAF1 UBX molecules in an asymmetric unit of the crystal show two different conformations of the FcisP touch-turn motif. The phenyl ring of F(619) in the motif stacks partly over cis-Pro(620) in one conformation, whereas it is swung out from cis-P(620), in the other conformation, and forms hydrophobic contacts with the residues of the neighboring molecule. In addition, the entire FcisP touch-turn motif is pulled out in the second conformation by about 2Å in comparison to the first conformation. Those conformational differences observed in the p97/VCP-binding motif caused by the interaction with neighboring molecules presumably represent the conformational change of the UBX domain on its binding to the N domain of p97/VCP.
Organizational Affiliation:
Department of Chemistry, College of Natural Sciences, Soongsil University, Seoul 156-743, Republic of Korea.