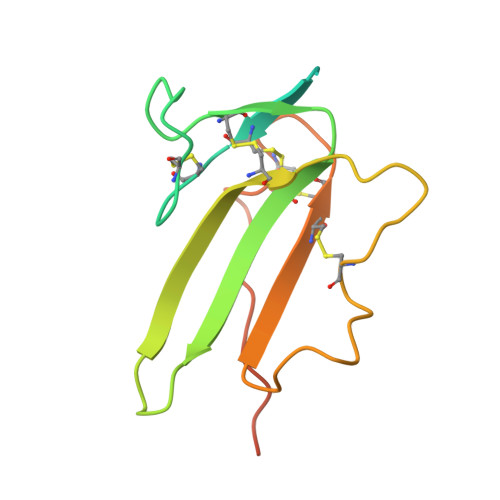
A selection fit mechanism in BMP receptor IA as a possible source for BMP ligand-receptor promiscuity
Harth, S., Kotzsch, A., Hu, J., Sebald, W., Mueller, T.D.(2010) PLoS One 5: e13049-e13049
- PubMed: 20927405
- DOI: https://doi.org/10.1371/journal.pone.0013049
- Primary Citation of Related Structures:
3NH7 - PubMed Abstract:
Members of the TGF-β superfamily are characterized by a highly promiscuous ligand-receptor interaction as is readily apparent from the numeral discrepancy of only seven type I and five type II receptors available for more than 40 ligands. Structural and functional studies have been used to address the question of how specific signals can be deduced from a limited number of receptor combinations and to unravel the molecular mechanisms underlying the protein-protein recognition that allow such limited specificity. In this study we have investigated how an antigen binding antibody fragment (Fab) raised against the extracellular domain of the BMP receptor type IA (BMPR-IA) recognizes the receptor's BMP-2 binding epitope and thereby neutralizes BMP-2 receptor activation. The crystal structure of the complex of the BMPR-IA ectodomain bound to the Fab AbD1556 revealed that the contact surface of BMPR-IA overlaps extensively with the contact surface for BMP-2 interaction. Although the structural epitopes of BMPR-IA to both binding partners coincides, the structures of BMPR-IA in the two complexes differ significantly. In contrast to the structural differences, alanine-scanning mutagenesis of BMPR-IA showed that the functional determinants for binding to the antibody and BMP-2 are almost identical. Comparing the structures of BMPR-IA bound to BMP-2 or bound to the Fab AbD1556 with the structure of unbound BMPR-IA shows that binding of BMPR-IA to its interaction partners follows a selection fit mechanism, possibly indicating that the ligand promiscuity of BMPR-IA is inherently encoded by structural adaptability. The functional and structural analysis of the BMPR-IA binding antibody AbD1556 mimicking the BMP-2 binding epitope may thus pave the way for the design of low-molecular weight synthetic receptor binders/inhibitors.
Organizational Affiliation:
Lehrstuhl für Physiologische Chemie II, Theodor-Boveri-Institut für Biowissenschaften der Universität Würzburg, Würzburg, Germany.