Solution Structure of Acidocin B, a Circular Bacteriocin Produced by Lactobacillus acidophilus M46.
Acedo, J.Z., van Belkum, M.J., Lohans, C.T., McKay, R.T., Miskolzie, M., Vederas, J.C.(2015) Appl Environ Microbiol 81: 2910-2918
- PubMed: 25681186
- DOI: https://doi.org/10.1128/AEM.04265-14
- Primary Citation of Related Structures:
2MWR - PubMed Abstract:
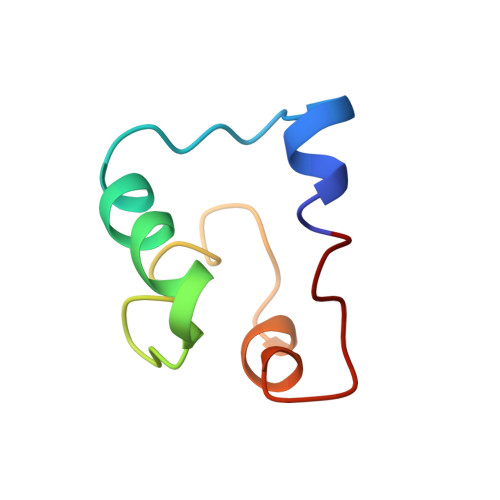
Acidocin B, a bacteriocin produced by Lactobacillus acidophilus M46, was originally reported to be a linear peptide composed of 59 amino acid residues. However, its high sequence similarity to gassericin A, a circular bacteriocin from Lactobacillus gasseri LA39, suggested that acidocin B might be circular as well. Acidocin B was purified from culture supernatant by a series of hydrophobic interaction chromatographic steps. Its circular nature was ascertained by matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry and tandem mass spectrometry (MS/MS) sequencing. The peptide sequence was found to consist of 58 amino acids with a molecular mass of 5,621.5 Da. The sequence of the acidocin B biosynthetic gene cluster was also determined and showed high nucleotide sequence similarity to that of gassericin A. The nuclear magnetic resonance (NMR) solution structure of acidocin B in sodium dodecyl sulfate micelles was elucidated, revealing that it is composed of four α-helices of similar length that are folded to form a compact, globular bundle with a central pore. This is a three-dimensional structure for a member of subgroup II circular bacteriocins, which are classified based on their isoelectric points of ∼7 or lower. Comparison of acidocin B with carnocyclin A, a subgroup I circular bacteriocin with four α-helices and a pI of 10, revealed differences in the overall folding. The observed variations could be attributed to inherent diversity in their physical properties, which also required the use of different solvent systems for three-dimensional structural elucidation.
Organizational Affiliation:
Department of Chemistry, University of Alberta, Edmonton, Alberta, Canada.