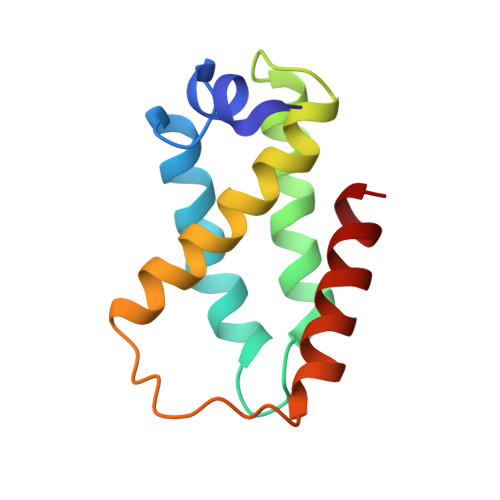
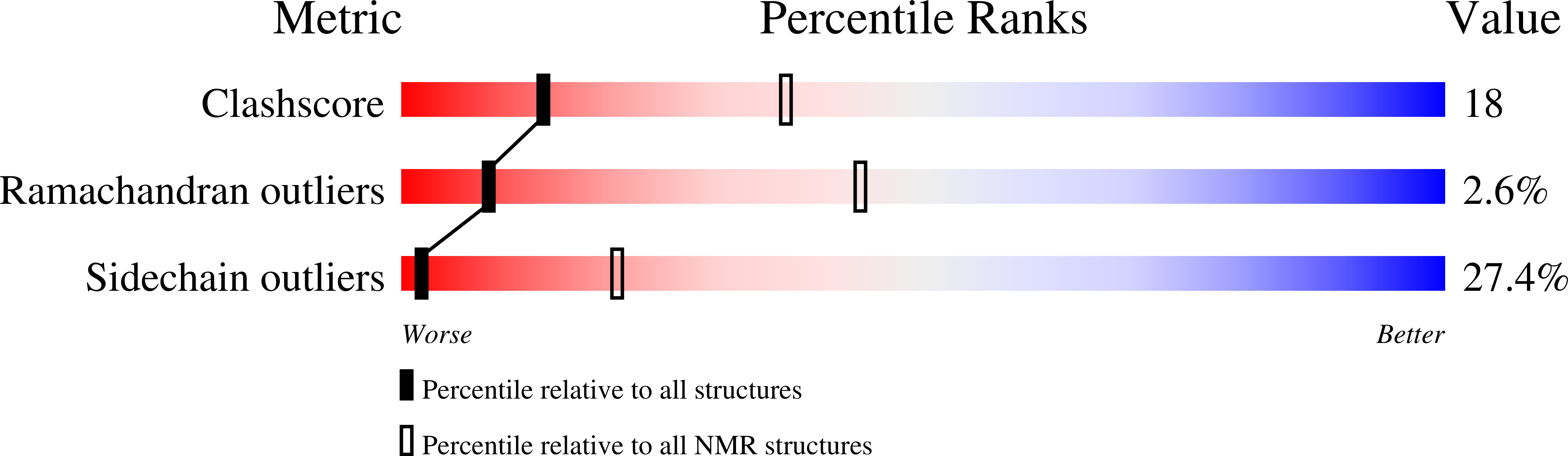Structure and function of C-terminal domain of aciniform spidroin
Wang, S., Huang, W., Yang, D.(2014) Biomacromolecules 15: 468-477
- PubMed: 24422432
- DOI: https://doi.org/10.1021/bm401709v
- Primary Citation of Related Structures:
2MAB - PubMed Abstract:
C-terminal domains (CTDs) of various spidroins are relatively conserved in the amino acid sequence and have been suggested to perform similar functions. Here, we solved the structure of the CTD of an aciniform spidroin using NMR spectroscopy with a two-point mutant and studied its functional role with several constructs. The CTDs of aciniform, major, and minor ampullate spidroins adopt the same domain-swapping dimeric folding although their sequence identities are 24-40%. Unlike CTDs of major and minor ampullate spidroins, the aciniform CTD had no obvious effects on preventing spidroins from aggregation in storage but slightly enhanced protein assembly under shear force. The differential functions may result from significant differences in detailed structures and properties of repetitive regions of various types of spidroins. Nevertheless, all CTDs may have a common functional role: dimerization of the CTD doubles the size of silk proteins, reduces the number of chain ends, and thus leads to fewer chain end defects in the fibers formed.
Organizational Affiliation:
Department of Biological Sciences, National University of Singapore , 14 Science Drive 4, Singapore 117543.