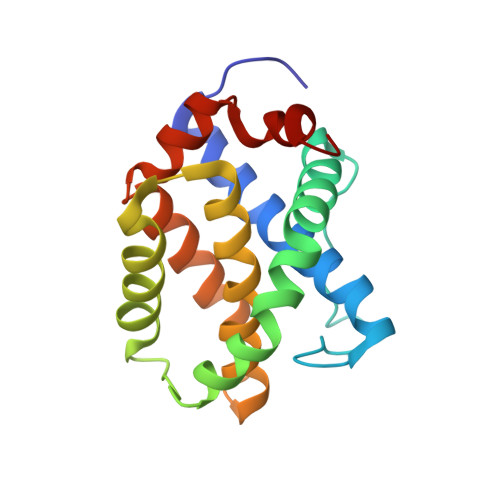
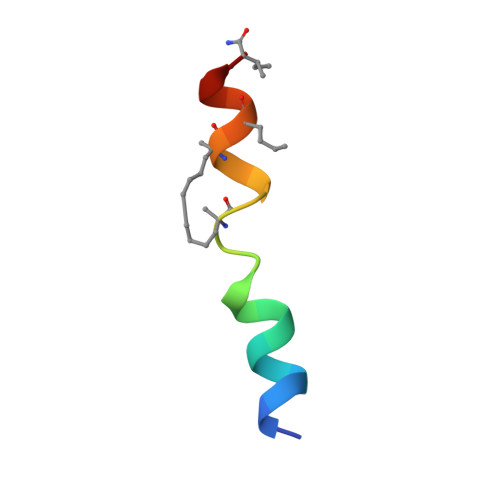
BID-induced structural changes in BAK promote apoptosis.
Moldoveanu, T., Grace, C.R., Llambi, F., Nourse, A., Fitzgerald, P., Gehring, K., Kriwacki, R.W., Green, D.R.(2013) Nat Struct Mol Biol 20: 589-597
- PubMed: 23604079
- DOI: https://doi.org/10.1038/nsmb.2563
- Primary Citation of Related Structures:
2M5B - PubMed Abstract:
The BCL-2-family protein BAK is responsible for mitochondrial outer-membrane permeabilization (MOMP), which leads to apoptosis. The BCL-2 homology 3 (BH3)-only protein BID activates BAK to perform this function. We report the NMR solution structure of the human BID BH3-BAK complex, which identified the activation site at the canonical BH3-binding groove of BAK. Mutating the BAK BH1 in the groove prevented activation and MOMP but not the binding of BID. BAK BH3 mutations allowed BID binding and activation but blunted function by blocking BAK oligomerization. BAK activation follows a 'hit-and-run' mechanism whereby BID dissociates from the trigger site, which allows BAK oligomerization at an overlapping interface. In contrast, the BH3-only proteins NOXA and BAD are predicted to clash with the trigger site and are not activators of BAK. These findings provide insights into the early stages of BAK activation.
Organizational Affiliation:
Department of Immunology, St. Jude Children's Research Hospital, Memphis, Tennessee, USA.