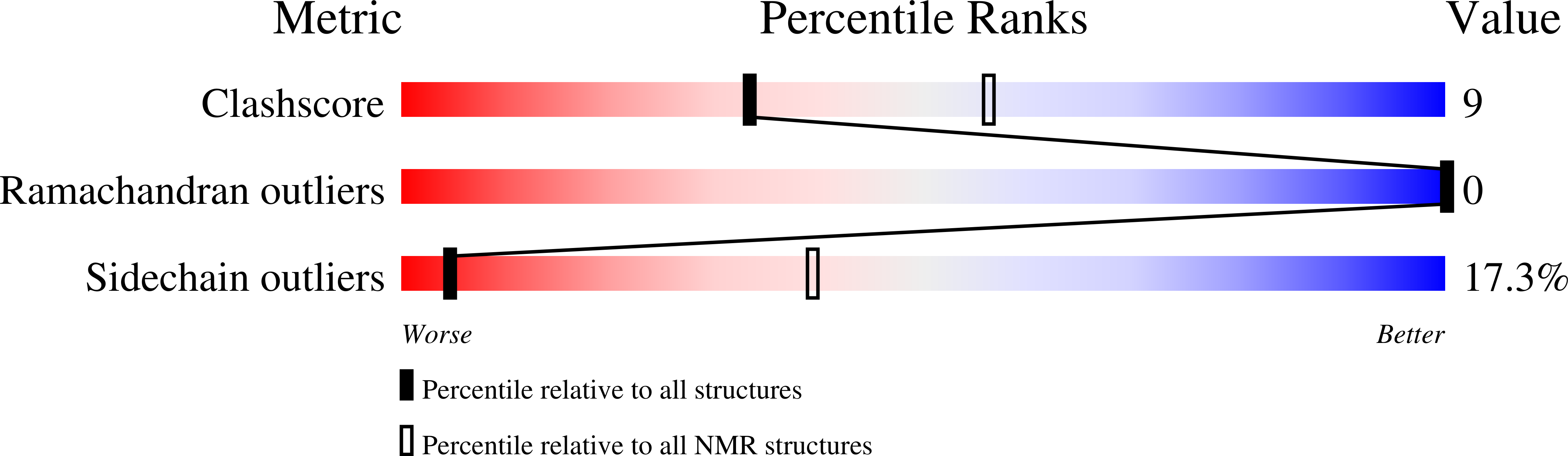Therapeutic potential of the Peptide leucine arginine as a new nonplant bowman-birk-like serine protease inhibitor.
Rothemund, S., Sonnichsen, F.D., Polte, T.(2013) J Med Chem 56: 6732-6744
- PubMed: 23988198
- DOI: https://doi.org/10.1021/jm4005362
- Primary Citation of Related Structures:
2M3N - PubMed Abstract:
The peptide leucine arginine (pLR) belongs to a new class of cyclic peptides isolated from frog skin. Its primary sequence is similar to the reactive loop of plant Bowman-Birk inhibitors (BBI), and the recently discovered circular sunflower trypsin inhibitor-1 (SFTI-1). The conformational properties of pLR in solution were determined by NMR spectroscopy and revealed excellent structural similarity to BBI and SFTI-1. Moreover, pLR is a highly potent trypsin inhibitor, with Ki values in the nanomolar range, and, due to its small size, a potential inhibitor of the serine protease tryptase. Since tryptase plays a crucial role in the development of allergic airway inflammation, the therapeutic potential of pLR in a murine asthma model was investigated. Treatment of ovalbumin-sensitized mice with pLR during allergen challenge reduced the acute asthma phenotype. Most importantly, application even at the end of a long-lasting chronic asthma model decreased the development of chronic airway inflammation and tissue remodeling.
Organizational Affiliation:
Interdisciplinary Center of Clinical Research, University of Leipzig , Leipzig, Germany.