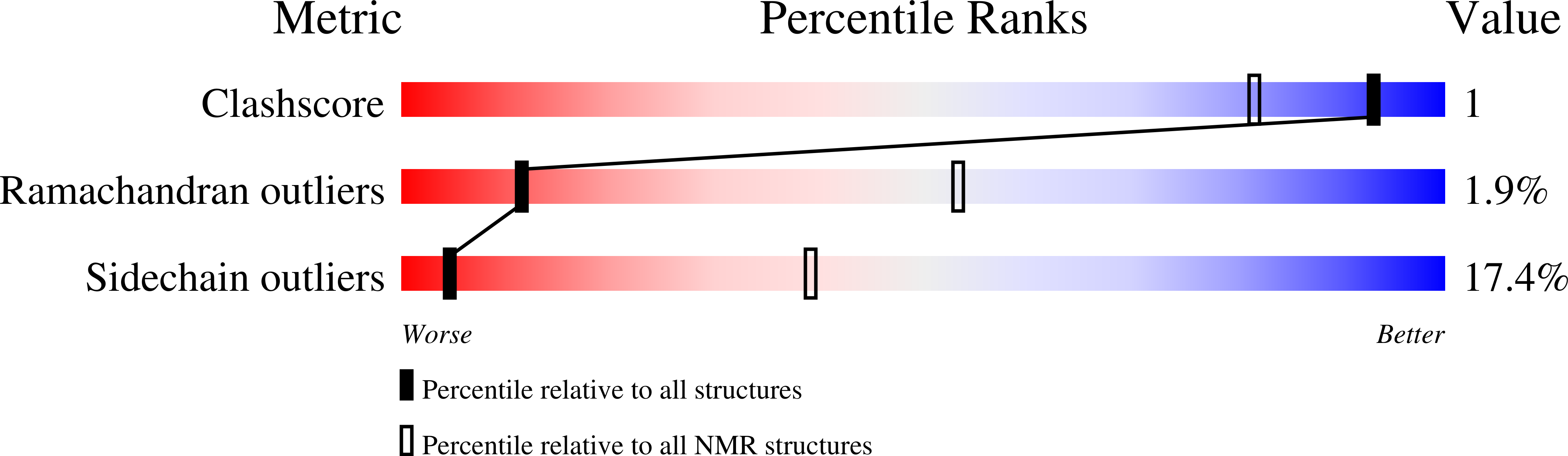Post-translational S-Nitrosylation Is an Endogenous Factor Fine Tuning the Properties of Human S100A1 Protein.
Lenarcic Zivkovic, M., Zareba-Koziol, M., Zhukova, L., Poznanski, J., Zhukov, I., Wyslouch-Cieszynska, A.(2012) J Biol Chem 287: 40457-40470
- PubMed: 22989881
- DOI: https://doi.org/10.1074/jbc.M112.418392
- Primary Citation of Related Structures:
2LLT, 2LLU - PubMed Abstract:
S100A1 protein is a proposed target of molecule-guided therapy for heart failure. S-Nitrosylation of S100A1 is present in cells, increases Ca(2+) binding, and tunes the overall protein conformation. Thiol-aromatic molecular switch is responsible for NO-related modification of S100A1 properties. Post-translational S-nitrosylation may provide functional diversity and specificity to S100A1 and other S100 protein family members. S100A1 is a member of the Ca(2+)-binding S100 protein family. It is expressed in brain and heart tissue, where it plays a crucial role as a modulator of Ca(2+) homeostasis, energy metabolism, neurotransmitter release, and contractile performance. Biological effects of S100A1 have been attributed to its direct interaction with a variety of target proteins. The (patho)physiological relevance of S100A1 makes it an important molecular target for future therapeutic intervention. S-Nitrosylation is a post-translational modification of proteins, which plays a role in cellular signal transduction under physiological and pathological conditions. In this study, we confirmed that S100A1 protein is endogenously modified by Cys(85) S-nitrosylation in PC12 cells, which are a well established model system for studying S100A1 function. We used isothermal calorimetry to show that S-nitrosylation facilitates the formation of Ca(2+)-loaded S100A1 at physiological ionic strength conditions. To establish the unique influence of the S-nitroso group, our study describes high resolution three-dimensional structures of human apo-S100A1 protein with the Cys(85) thiol group in reduced and S-nitrosylated states. Solution structures of the proteins are based on NMR data obtained at physiological ionic strength. Comparative analysis shows that S-nitrosylation fine tunes the overall architecture of S100A1 protein. Although the typical S100 protein intersubunit four-helix bundle is conserved upon S-nitrosylation, the conformation of S100A1 protein is reorganized at the sites most important for target recognition (i.e. the C-terminal helix and the linker connecting two EF-hand domains). In summary, this study discloses cysteine S-nitrosylation as a new factor responsible for increasing functional diversity of S100A1 and helps explain the role of S100A1 as a Ca(2+) signal transmitter sensitive to NO/redox equilibrium within cells.
Organizational Affiliation:
National Institute of Chemistry, Slovenian NMR Centre, Hajdrihova 19, 1001 Ljubljana, Slovenia.