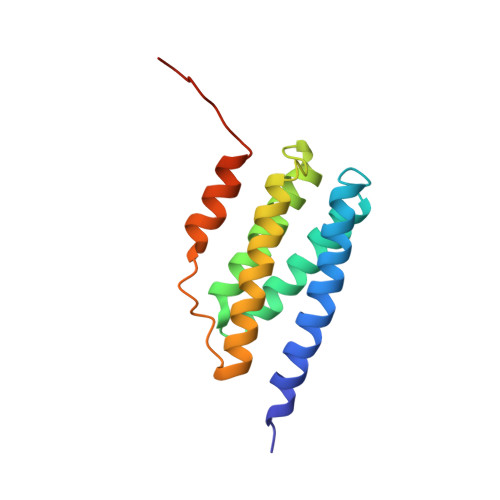
The structure and dynamic properties of the complete histidine phosphotransfer domain of the chemotaxis specific histidine autokinase CheA from Thermotoga maritima.
Vu, A., Hamel, D.J., Zhou, H., Dahlquist, F.W.(2011) J Biomol NMR 51: 49-55
- PubMed: 21947914
- DOI: https://doi.org/10.1007/s10858-011-9540-2
- Primary Citation of Related Structures:
2LD6 - PubMed Abstract:
The bacterial histidine autokinase CheA contains a histidine phosphotransfer (Hpt) domain that accepts a phosphate from the catalytic domain and donates the phosphate to either target response regulator protein, CheY or CheB. The Hpt domain forms a helix-bundle structure with a conserved four-helix bundle motif and a variable fifth helix. Observation of two nearly equally populated conformations in the crystal structure of a Hpt domain fragment of CheA from Thermotoga maritima containing only the first four helices suggests more mobility in a tightly packed helix bundle structure than previously thought. In order to examine how the structures of Hpt domain homologs may differ from each other particularly in the conformation of the last helix, and whether an alternative conformation exists in the intact Hpt domain in solution, we have solved a high-resolution, solution structure of the CheA Hpt from T. maritima and characterized the backbone dynamics of this protein. The structure contains a four-helix bundle characteristic of histidine phosphotransfer domains. The position and orientation of the fifth helix resembles those in known Hpt domain crystal and solution structures in other histidine kinases. The alternative conformation that was reported in the crystal structure of the CheA Hpt from T. maritima missing the fifth helix is not detected in the solution structure, suggesting a role for the fifth helix in providing stabilizing forces to the overall structure.
Organizational Affiliation:
Department of Chemistry and Biochemistry, University of California Santa Barbara, Santa Barbara, CA 93103, USA.