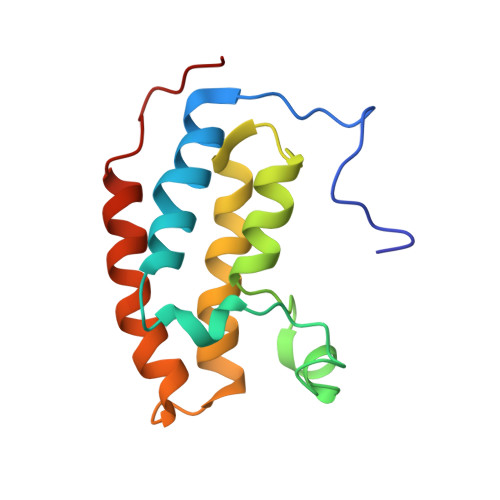
Structural basis and specificity of acetylated transcription factor GATA1 recognition by BET family bromodomain protein Brd3.
Gamsjaeger, R., Webb, S.R., Lamonica, J.M., Billin, A., Blobel, G.A., Mackay, J.P.(2011) Mol Cell Biol 31: 2632-2640
- PubMed: 21555453
- DOI: https://doi.org/10.1128/MCB.05413-11
- Primary Citation of Related Structures:
2L5E - PubMed Abstract:
Recent data demonstrate that small synthetic compounds specifically targeting bromodomain proteins can modulate the expression of cancer-related or inflammatory genes. Although these studies have focused on the ability of bromodomains to recognize acetylated histones, it is increasingly becoming clear that histone-like modifications exist on other important proteins, such as transcription factors. However, our understanding of the molecular mechanisms through which these modifications modulate protein function is far from complete. The transcription factor GATA1 can be acetylated at lysine residues adjacent to the zinc finger domains, and this acetylation is essential for the normal chromatin occupancy of GATA1. We have recently identified the bromodomain-containing protein Brd3 as a cofactor that interacts with acetylated GATA1 and shown that this interaction is essential for the targeting of GATA1 to chromatin. Here we describe the structural basis for this interaction. Our data reveal for the first time the molecular details of an interaction between a transcription factor bearing multiple acetylation modifications and its cognate recognition module. We also show that this interaction can be inhibited by an acetyllysine mimic, highlighting the importance of further increasing the specificity of compounds that target bromodomain and extraterminal (BET) bromodomains in order to fully realize their therapeutic potential.
Organizational Affiliation:
School of Molecular Bioscience, University of Sydney, Sydney, NSW 2006, Australia.