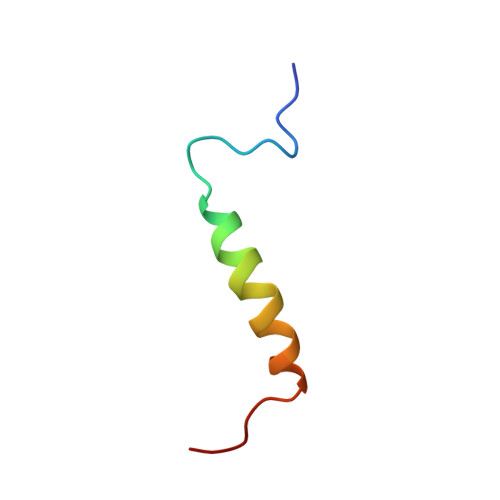
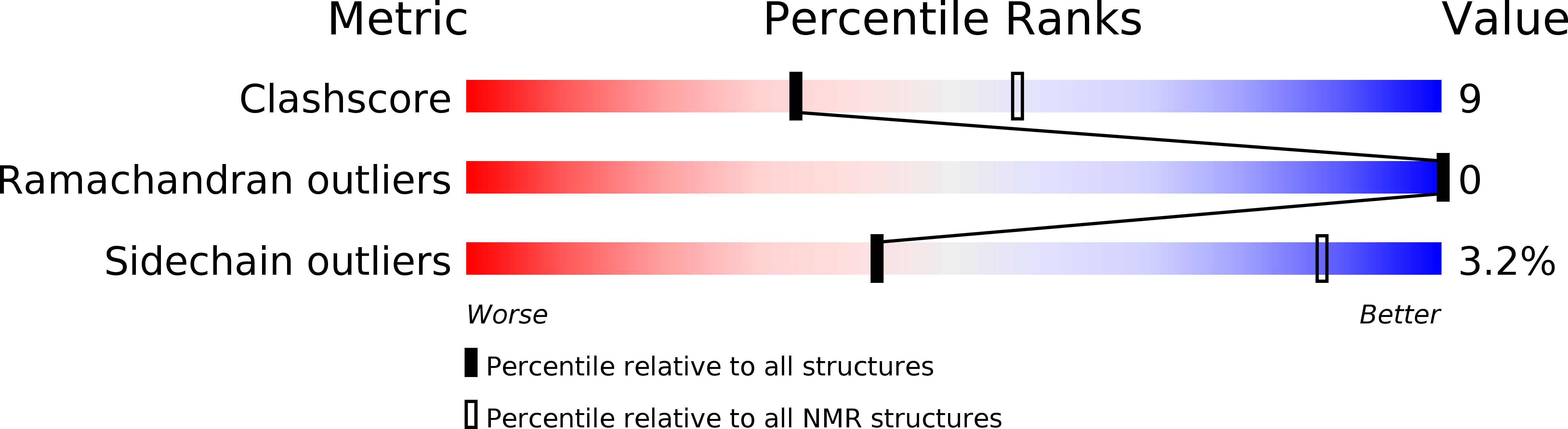Solution Structure of the Leader Sequence of the Patellamide Precursor Peptide, PatE(1-34).
Houssen, W.E., Wright, S.H., Kalverda, A.P., Thompson, G.S., Kelly, S.M., Jaspars, M.(2010) Chembiochem 11: 1867-1873
- PubMed: 20715266
- DOI: https://doi.org/10.1002/cbic.201000305
- Primary Citation of Related Structures:
2KYA - PubMed Abstract:
The solution structure of the leader sequence of the patellamide precursor peptide was analysed by using CD and determined with NOE-restrained molecular dynamics calculations. This leader sequence is highly conserved in the precursor peptides of some other cyanobactins harbouring heterocycles, and is assumed to play a role in targeting the precursor peptide to the post-translational machinery. The sequence was observed to form an alpha-helix spanning residues 13-28 with a hydrophobic surface on one side of the helix. This hydrophobic surface is proposed to be the site of the initial binding with modifying enzymes.
Organizational Affiliation:
Marine Biodiscovery Centre, Department of Chemistry, University of Aberdeen, Aberdeen AB24 3UE, Scotland, UK.