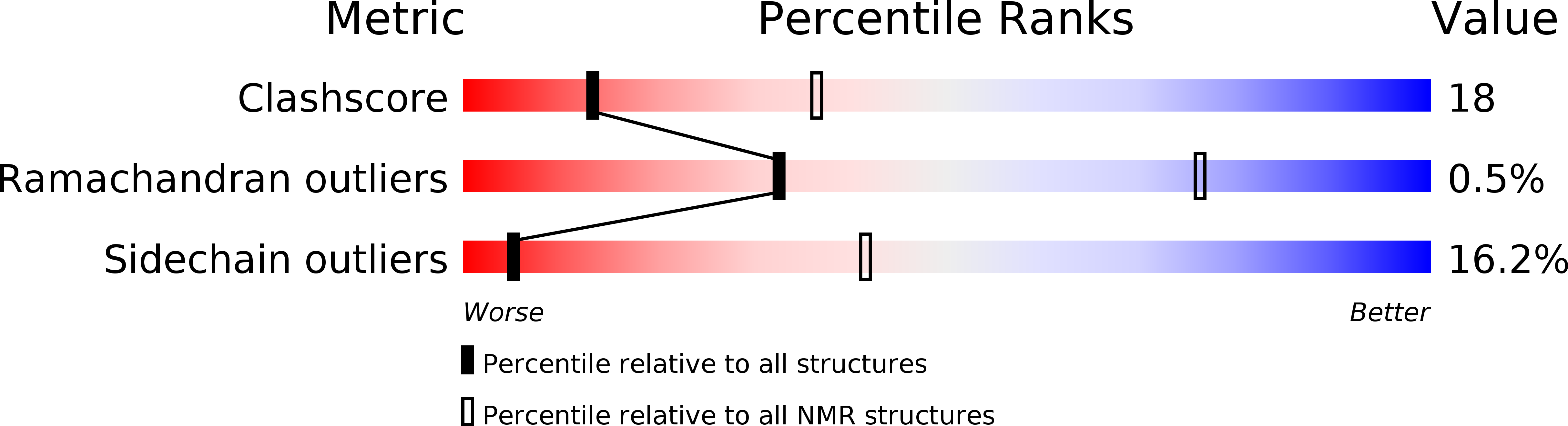Biochemical properties and catalytic domain structure of the CcmH protein from Escherichia coli.
Zheng, X.M., Hong, J., Li, H.Y., Lin, D.H., Hu, H.Y.(2012) Biochim Biophys Acta 1824: 1394-1400
- PubMed: 22789558
- DOI: https://doi.org/10.1016/j.bbapap.2012.06.017
- Primary Citation of Related Structures:
2KW0 - PubMed Abstract:
In the Gram-negative bacterium of Escherichia coli, eight genes organized as a ccm operon (ccmABCDEFGH) are involved in the maturation of c-type cytochromes. The proteins encoded by the last three genes ccmFGH are believed to form a lyase complex functioning in the reduction of apocytochrome c and haem attachment. Among them, CcmH is a membrane-associated protein; its N-terminus is a catalytic domain with the active CXXC motif and the C-terminus is predicted as a TPR-like domain with unknown function. By using SCAM (scanning cysteine accessibility mutagenesis) and Gaussia luciferase fusion assays, we provide experimental evidence for the entire topological structure of E. coli CcmH. The mature CcmH is a periplasm-resident oxidoreductase anchored to the inner membrane by two transmembrane segments. Both N- and C-terminal domains are located and function in the periplasmic compartment. Moreover, the N-terminal domain forms a monomer in solution, while the C-terminal domain is a compact fold with helical structures. The NMR solution structure of the catalytic domain in reduced form exhibits mainly a three-helix bundle, providing further information for the redox mechanism. The redox potential suggests that CcmH exhibits a strong reductase that may function in the last step of reduction of apocytochrome c for haem attachment.
Organizational Affiliation:
Institute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, China.