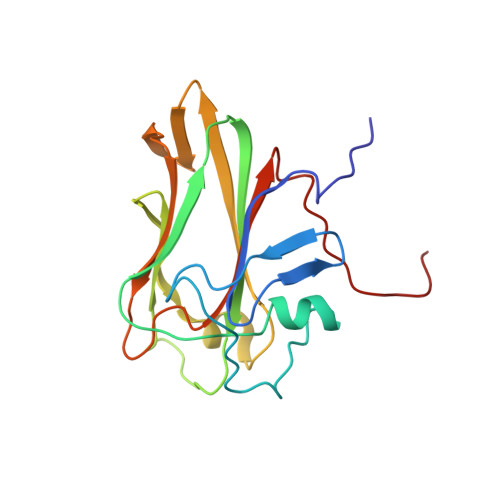
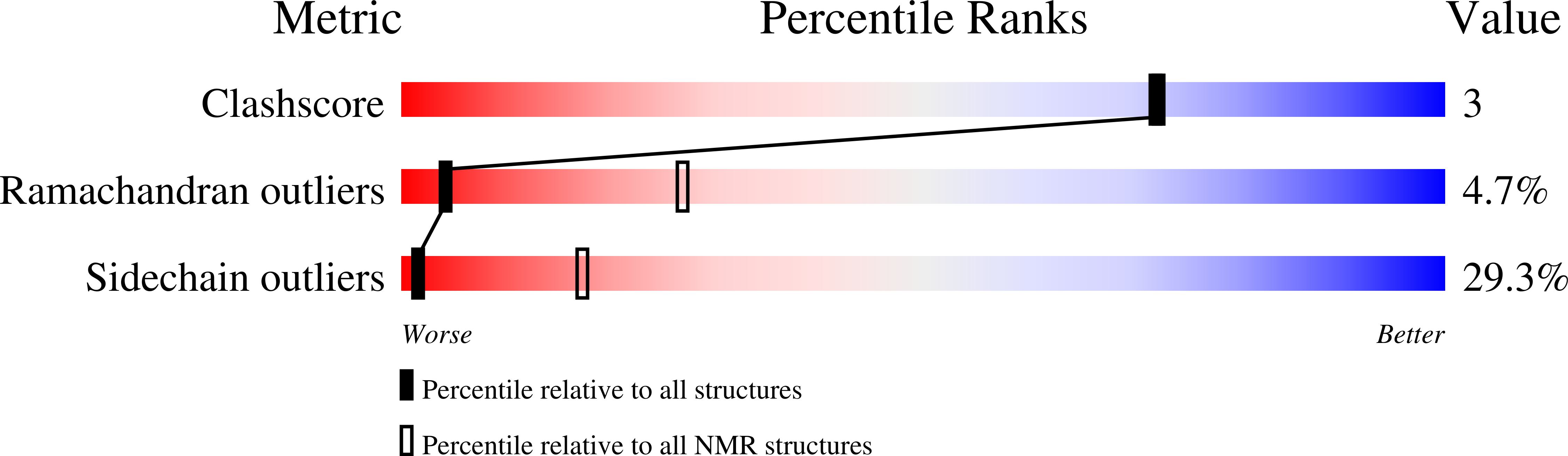Analysis of the specific interactions between the lectin domain of malectin and diglucosides.
Schallus, T., Feher, K., Sternberg, U., Rybin, V., Muhle-Goll, C.(2010) Glycobiology 20: 1010-1020
- PubMed: 20466650
- DOI: https://doi.org/10.1093/glycob/cwq059
- Primary Citation of Related Structures:
2KR2 - PubMed Abstract:
The endoplasmic reticulum malectin is a highly conserved protein in the animal kingdom that has no counterpart so far in lower organisms. We recently determined the structure of its conserved domain and found a highly selective binding to Glc(2)Man(9)GlcNAc(2), an intermediate of N-glycosylation. In our quest for putative ligands during the initial characterization of the protein, we noticed that the malectin domain is highly specific for diglucosides but quite tolerant towards the linkage of the glucosidic bond. To understand the molecular requirements for the observed promiscuity of the malectin domain, here we analyze the binding to a range of diglucosides through comparison of the protein chemical shift perturbation patterns and the saturation transfer difference spectra of the ligands including two maltose-mimicking drugs. A comparison of the maltose-bound structure of the malectin domain with the complex of the native ligand nigerose reveals why malectin is able to tolerate such a diversity of ligands.
Organizational Affiliation:
EMBL, Meyerhofstrasse 1, 69117 Heidelberg, Germany.