NMR structure of Clostridium perfringens protein CPE0013.
Ding, K., Ramelot, T.A., Wang, H., Nwosu, C., Montelione, G.T., Kennedy, M.A.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Hypothetical protein CPE0013 | 85 | Clostridium perfringens | Mutation(s): 0 | 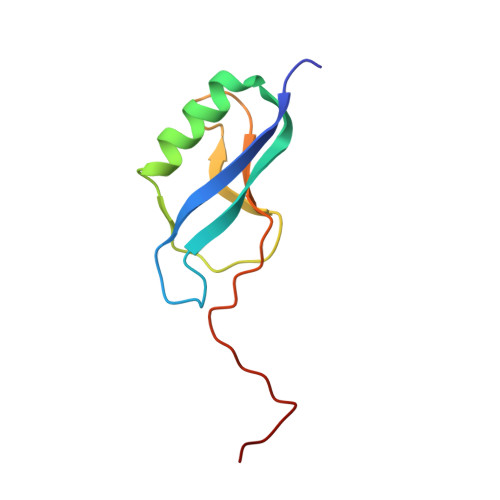 | |
UniProt | |||||
Find proteins for Q8XPF0 (Clostridium perfringens (strain 13 / Type A)) Explore Q8XPF0 Go to UniProtKB: Q8XPF0 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q8XPF0 | ||||
Sequence AnnotationsExpand | |||||
| |||||