NMR Solution Structure, Stability, and Interaction of the Recombinant Bovine Fibrinogen alphaC-Domain Fragment
Burton, R.A., Tsurupa, G., Hantgan, R.R., Tjandra, N., Medved, L.(2007) Biochemistry 46: 8550-8560
- PubMed: 17590019
- DOI: https://doi.org/10.1021/bi700606v
- Primary Citation of Related Structures:
2JOR - PubMed Abstract:
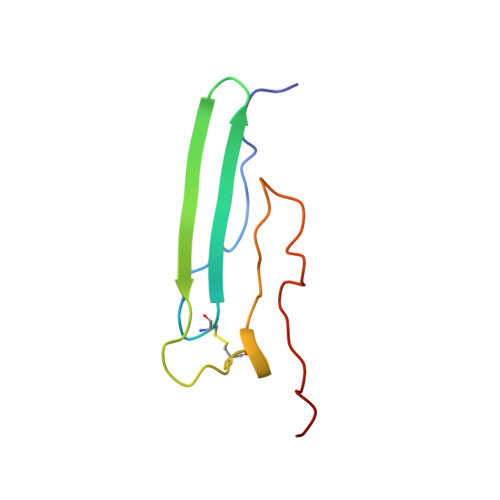
According to the existing hypothesis, in fibrinogen, the COOH-terminal portions of two Aalpha chains are folded into compact alphaC-domains that interact intramolecularly with each other and with the central region of the molecule; in fibrin, the alphaC-domains switch to an intermolecular interaction resulting in alphaC-polymers. In agreement, our recent NMR study identified within the bovine fibrinogen Aalpha374-538 alphaC-domain fragment an ordered compact structure including a beta-hairpin restricted at the base by a 423-453 disulfide linkage. To establish the complete structure of the alphaC-domain and to further test the hypothesis, we expressed a shorter alphaC-fragment, Aalpha406-483, and performed detailed analysis of its structure, stability, and interactions. NMR experiments on the Aalpha406-483 fragment identified a second loose beta-hairpin formed by residues 459-476, yielding a structure consisting of an intrinsically unstable mixed parallel/antiparallel beta-sheet. Size-exclusion chromatography and sedimentation velocity experiments revealed that the Aalpha406-483 fragment forms soluble oligomers whose fraction increases with an increase in concentration. This was confirmed by sedimentation equilibrium analysis, which also revealed that the addition of each monomer to an assembling alphaC-oligomer substantially increases its stabilizing free energy. In agreement, unfolding experiments monitored by CD established that oligomerization of Aalpha406-483 results in increased thermal stability. Altogether, these experiments establish the complete NMR solution structure of the Aalpha406-483 alphaC-domain fragment, provide direct evidence for the intra- and intermolecular interactions between the alphaC-domains, and confirm that these interactions are thermodynamically driven.
Organizational Affiliation:
Laboratory of Molecular Biophysics, National Heart, Lung, and Blood Institute, National Institutes of Health, 50 Center Drive, Bethesda, Maryland 20892, USA.