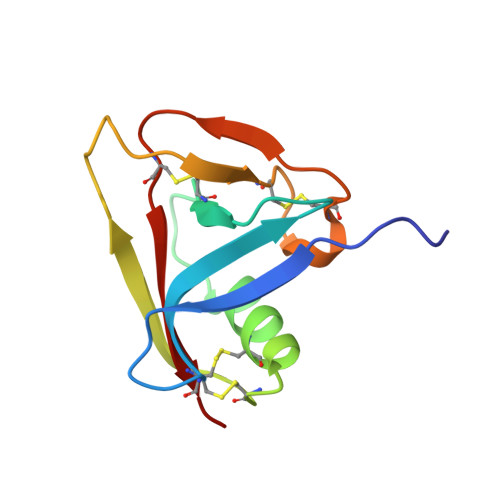
Crystal Structure of the Third Extracellular Domain of Cd5 Reveals the Fold of a Group B Scavenger Cysteine-Rich Receptor Domain.
Rodamilans, B., Munoz, I.G., Bragado-Nilsson, E., Sarrias, M.R., Padilla, O., Blanco, F.J., Lozano, F., Montoya, G.(2007) J Biol Chem 282: 12669
- PubMed: 17322294
- DOI: https://doi.org/10.1074/jbc.M611699200
- Primary Citation of Related Structures:
2JA4, 2OTT - PubMed Abstract:
Scavenger receptor cysteine-rich (SRCR) domains are ancient protein modules widely found among cell surface and secreted proteins of the innate and adaptive immune system, where they mediate ligand binding. We have solved the crystal structure at 2.2 A of resolution of the SRCR CD5 domain III, a human lymphocyte receptor involved in the modulation of antigen specific receptor-mediated T cell activation and differentiation signals. The first structure of a member of a group B SRCR domain reveals the fold of this ancient protein module into a central core formed by two antiparallel beta-sheets and one alpha-helix, illustrating the conserved core at the protein level of genes coding for group A and B members of the SRCR superfamily. The novel SRCR group B structure permits the interpretation of site-directed mutagenesis data on the binding of activated leukocyte cell adhesion molecule (ALCAM/CD166) binding to CD6, a closely related lymphocyte receptor homologue to CD5.
Organizational Affiliation:
Spanish National Cancer Center (CNIO), Structural Biology and Biocomputing Program, c/Melchor Fernandez Almagro 3, 28029 Madrid, Spain.