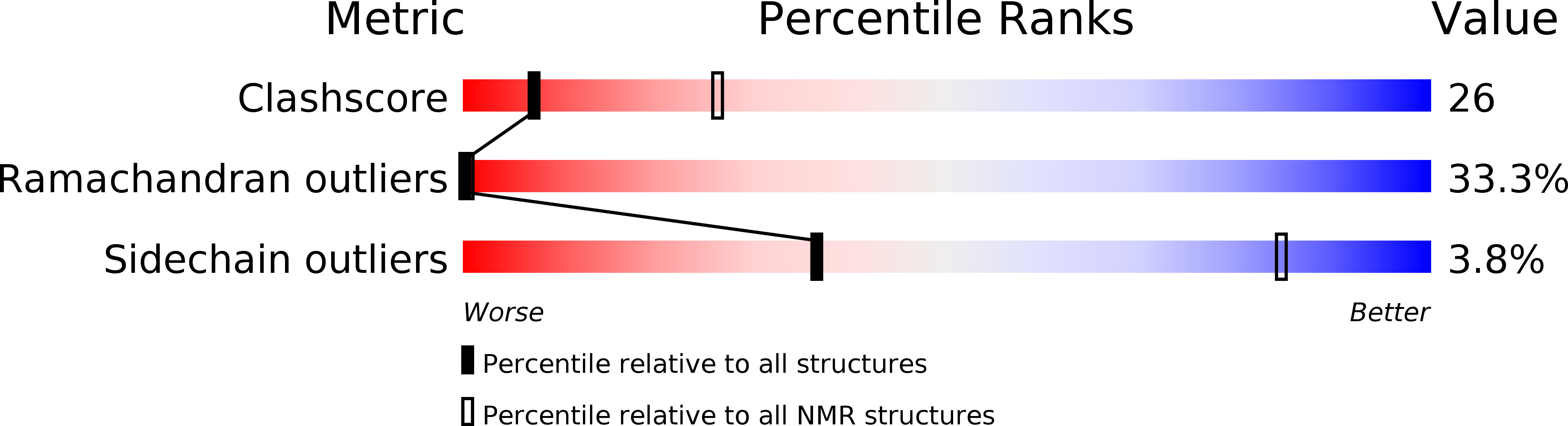Structure-function analysis of tritrpticin analogs: potential relationships between antimicrobial activities, model membrane interactions, and their micelle-bound NMR structures
Schibli, D.J., Nguyen, L.T., Kernaghan, S.D., Rekdal, O., Vogel, H.J.(2006) Biophys J 91: 4413-4426
- PubMed: 16997878
- DOI: https://doi.org/10.1529/biophysj.106.085837
- Primary Citation of Related Structures:
2I1D, 2I1E, 2I1F, 2I1G, 2I1H, 2I1I - PubMed Abstract:
Tritrpticin is a member of the cathelicidin family of antimicrobial peptides. Starting from its native sequence (VRRFPWWWPFLRR), eight synthetic peptide analogs were studied to investigate the roles of specific residues in its biological and structural properties. This included amidation of the C-terminus paired with substitutions of its cationic and Phe residues, as well as the Pro residues that are important for its two-turn micelle-bound structure. These analogs were determined to have a significant antimicrobial potency. In contrast, two other peptide analogs, those with the three Trp residues substituted with either Phe or Tyr residues are not highly membrane perturbing, as determined by leakage and flip-flop assays using fluorescence spectroscopy. Nevertheless the Phe analog has a high activity; this suggests an intracellular mechanism for antimicrobial activity that may be part of the overall mechanism of action of native tritrpticin as a complement to membrane perturbation. NMR experiments of these two Trp-substituted peptides showed the presence of multiple conformers. The structures of the six remaining Trp-containing analogs bound to dodecylphosphocholine micelles showed major, well-defined conformations. These peptides are membrane disruptive and show a wide range in hemolytic activity. Their micelle-bound structures either retain the typical turn-turn structure of native tritrpticin or have an extended alpha-helix. This work demonstrates that closely related antimicrobial peptides can often have remarkably altered properties with complex influences on their biological activities.
Organizational Affiliation:
Structural Biology Research Group, Department of Biological Sciences, University of Calgary, Calgary, Alberta, Canada.