X-ray snap shots of Inhibitor Binding Process in Photo-reactive Nitrile Hydratase
Nojiri, M., Kawano, Y., Hashimoto, K., Kamiya, N.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Nitrile hydratase subunit alpha | 206 | Rhodococcus erythropolis | Mutation(s): 1 EC: 4.2.1.84 | 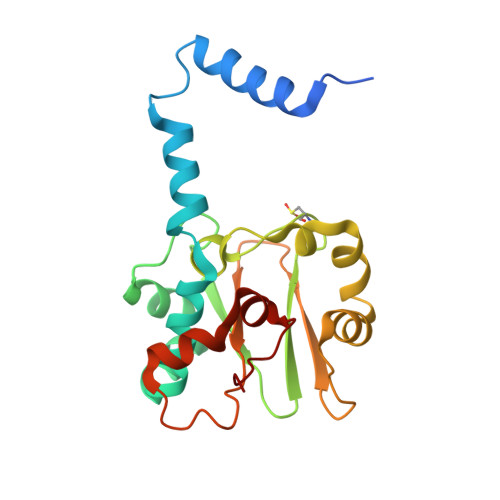 | |
UniProt | |||||
Find proteins for P13448 (Rhodococcus erythropolis) Explore P13448 Go to UniProtKB: P13448 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P13448 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Nitrile hydratase subunit beta | 212 | Rhodococcus erythropolis | Mutation(s): 0 EC: 4.2.1.84 | 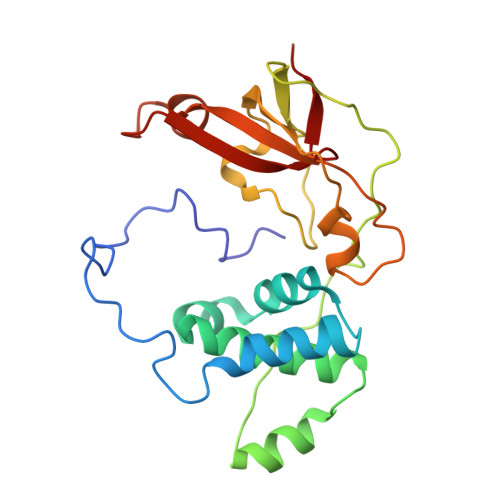 | |
UniProt | |||||
Find proteins for P13449 (Rhodococcus erythropolis) Explore P13449 Go to UniProtKB: P13449 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P13449 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| CYI Query on CYI | H [auth A] | CYCLOHEXYL ISOCYANIDE C7 H11 N XYZMOVWWVXBHDP-UHFFFAOYSA-N |  | ||
| FE Query on FE | C [auth A] | FE (III) ION Fe VTLYFUHAOXGGBS-UHFFFAOYSA-N |  | ||
| MG Query on MG | D [auth A] E [auth A] F [auth A] G [auth A] I [auth B] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| CSD Query on CSD | A | L-PEPTIDE LINKING | C3 H7 N O4 S |  | CYS |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 114.352 | α = 90 |
| b = 60.201 | β = 125.1 |
| c = 81.585 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| HKL-2000 | data reduction |
| SCALEPACK | data scaling |
| CNS | phasing |