High-resolution crystal structure of a truncated ColE7 translocation domain: implications for colicin transport across membranes
Cheng, Y.S., Shi, Z., Doudeva, L.G., Yang, W.Z., Chak, K.F., Yuan, H.S.(2006) J Mol Biol 356: 22-31
- PubMed: 16360169
- DOI: https://doi.org/10.1016/j.jmb.2005.11.056
- Primary Citation of Related Structures:
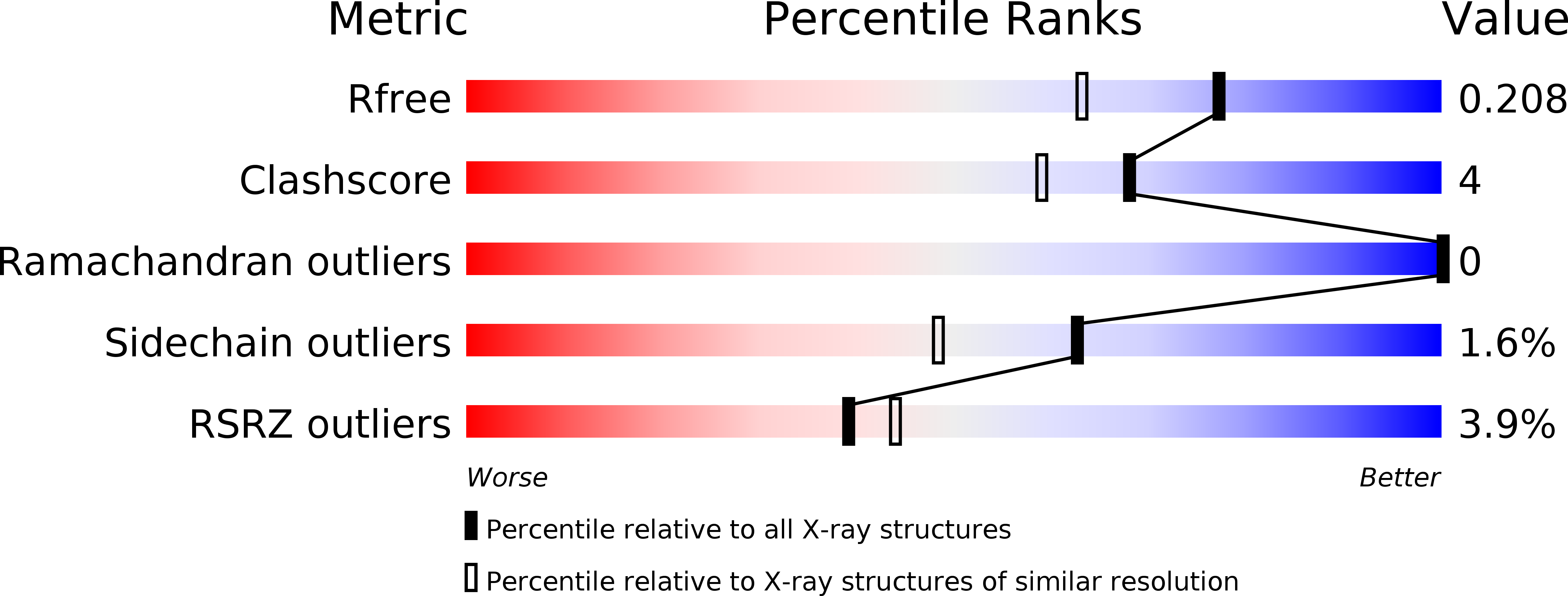
2AXC - PubMed Abstract:
ColE7 is a nuclease-type colicin released from Escherichia coli to kill sensitive bacterial cells by degrading the nucleic acid molecules in their cytoplasm. ColE7 is classified as one of the group A colicins, since the N-terminal translocation domain (T-domain) of the nuclease-type colicins interact with specific membrane-bound or periplasmic Tol proteins during protein import. Here, we show that if the N-terminal tail of ColE7 is deleted, ColE7 (residues 63-576) loses its bactericidal activity against E.coli. Moreover, TolB protein interacts directly with the T-domain of ColE7 (residues 1-316), but not with the N-terminal deleted T-domain (residues 60-316), as detected by co-immunoprecipitation experiments, confirming that the N-terminal tail is required for ColE7 interactions with TolB. The crystal structure of the N-terminal tail deleted ColE7 T-domain was determined by the multi-wavelength anomalous dispersion method at a resolution of 1.7 angstroms. The structure of the ColE7 T-domain superimposes well with the T-domain of ColE3 and TR-domain of ColB, a group A Tol-dependent colicin and a group B TonB-dependent colicin, respectively. The structural resemblance of group A and B colicins implies that the two groups of colicins may share a mechanistic connection during cellular import.
Organizational Affiliation:
Institute of Molecular Biology, Academia Sinica, Taipei, Taiwan, ROC.