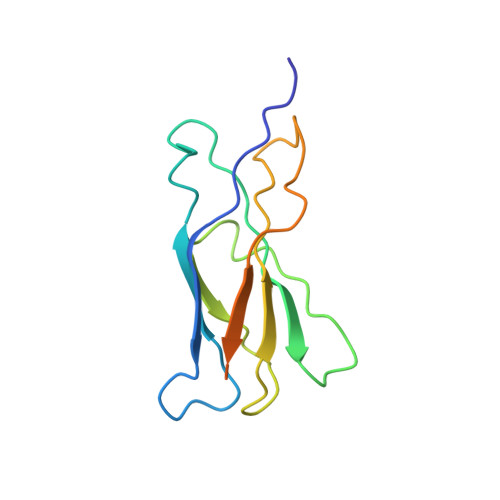
The solution structure of the membrane-proximal cytokine receptor domain of the human interleukin-6 receptor
Hecht, O., Dingley, A.J., Schwanter, A., Ozbek, S., Rose-John, S., Grotzinger, J.(2006) Biol Chem 387: 1255-1259
- PubMed: 16972794
- DOI: https://doi.org/10.1515/BC.2006.155
- Primary Citation of Related Structures:
2ARW - PubMed Abstract:
The members of the interleukin-6-type family of cytokines interact with receptors that have a modular structure and are built of several immunoglobulin-like and fibronectin type III-like domains. These receptors have a characteristic cytokine receptor homology region consisting of two fibronectin type III-like domains defined by a set of four conserved cysteines and a tryptophan-serine-X-tryptophan-serine sequence motif. On target cells, interleukin-6 (IL-6) initially binds to its cognate alpha-receptor and subsequently to a homodimer of the signal transducer receptor gp130. The IL-6 receptor (IL-6R) consists of three extracellular domains. The N-terminal immunoglobulin-like domain is not involved in ligand binding, whereas the third membrane-proximal fibronectin-like domain (IL-6R-D3) accounts for more than 90% of the binding energy to IL-6. Here, we present the solution structure of the IL-6R-D3 domain solved by multidimensional heteronuclear NMR spectroscopy.
Organizational Affiliation:
Biochemisches Institut der Christian-Albrechts-Universität Kiel, Olshausenstr. 40, D-24118 Kiel, Germany.