Structure and Ligand Selection of Hemoglobin II from Lucina pectinata
Gavira, J.A., Camara-Artigas, A., De Jesus-Bonilla, W., Lopez-Garriga, J., Lewis, A., Pietri, R., Yeh, S.R., Cadilla, C.L., Garcia-Ruiz, J.M.(2008) J Biol Chem 283: 9414-9423
- PubMed: 18203714
- DOI: https://doi.org/10.1074/jbc.M705026200
- Primary Citation of Related Structures:
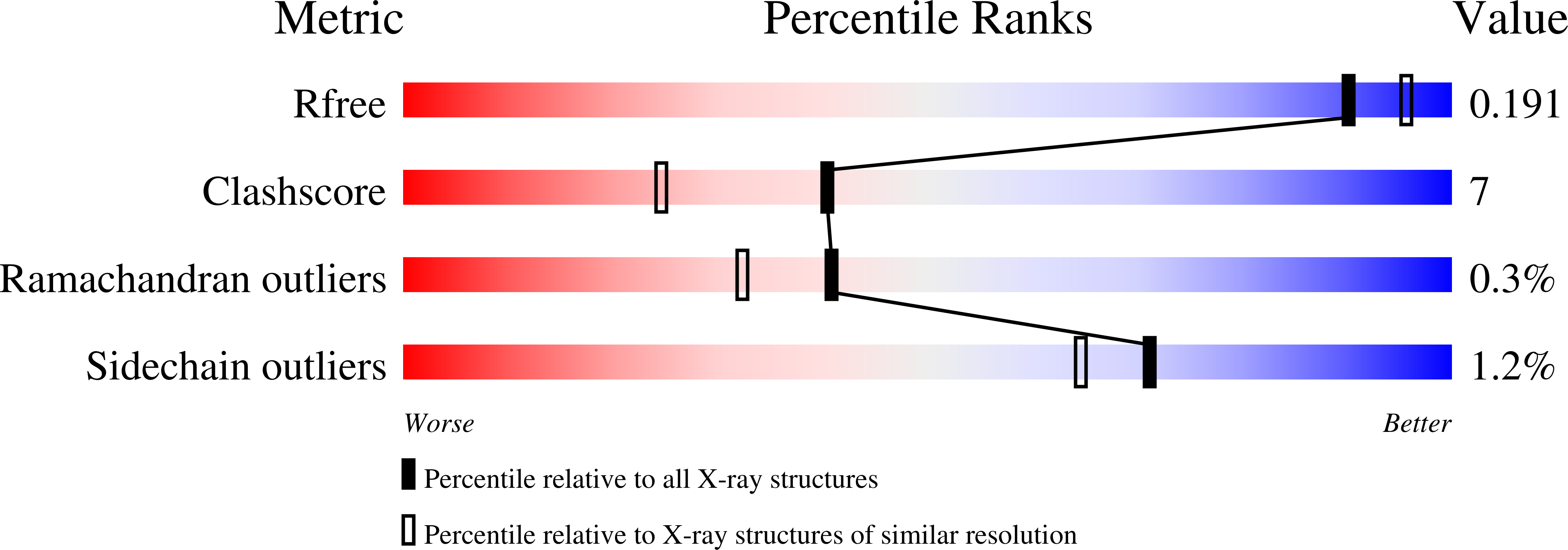
2OLP - PubMed Abstract:
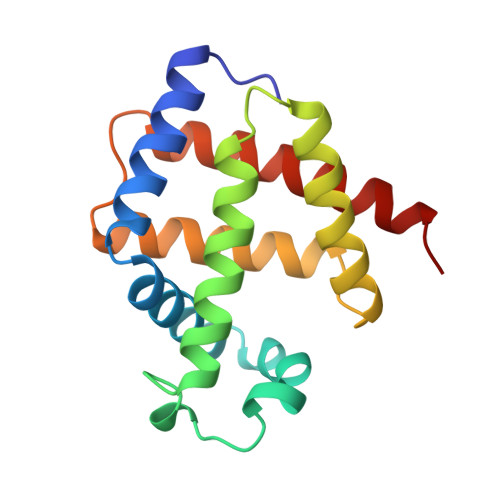
Lucina pectinata ctenidia harbor three heme proteins: sulfide-reactive hemoglobin I (HbI(Lp)) and the oxygen transporting hemoglobins II and III (HbII(Lp) and HbIII(Lp)) that remain unaffected by the presence of H(2)S. The mechanisms used by these three proteins for their function, including ligand control, remain unknown. The crystal structure of oxygen-bound HbII(Lp) shows a dimeric oxyHbII(Lp) where oxygen is tightly anchored to the heme through hydrogen bonds with Tyr(30)(B10) and Gln(65)(E7). The heme group is buried farther within HbII(Lp) than in HbI(Lp). The proximal His(97)(F8) is hydrogen bonded to a water molecule, which interacts electrostatically with a propionate group, resulting in a Fe-His vibration at 211 cm(-1). The combined effects of the HbII(Lp) small heme pocket, the hydrogen bonding network, the His(97) trans-effect, and the orientation of the oxygen molecule confer stability to the oxy-HbII(Lp) complex. Oxidation of HbI(Lp) Phe(B10) --> Tyr and HbII(Lp) only occurs when the pH is decreased from pH 7.5 to 5.0. Structural and resonance Raman spectroscopy studies suggest that HbII(Lp) oxygen binding and transport to the host bacteria may be regulated by the dynamic displacements of the Gln(65)(E7) and Tyr(30)(B10) pair toward the heme to protect it from changes in the heme oxidation state from Fe(II) to Fe(III).
Organizational Affiliation:
Laboratorio de Estudios Cristalográficos, CSIC, P.T. Ciencias de la Salud, Granada, Spain.