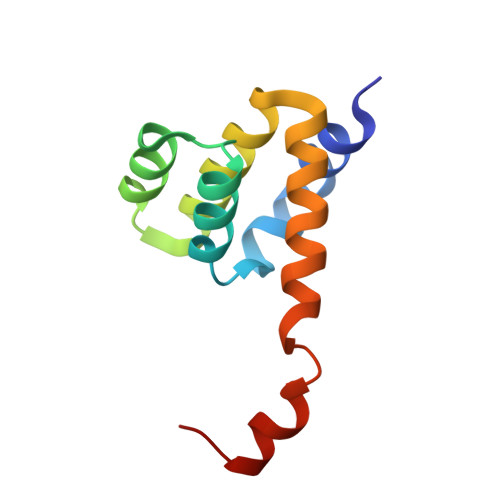
Crystal structure of the Nod1 caspase activation and recruitment domain.
Coussens, N.P., Mowers, J.C., McDonald, C., Nunez, G., Ramaswamy, S.(2007) Biochem Biophys Res Commun 353: 1-5
- PubMed: 17173864
- DOI: https://doi.org/10.1016/j.bbrc.2006.11.122
- Primary Citation of Related Structures:
2NSN - PubMed Abstract:
Nod-like receptors (NLRs), Nod1 and Nod2 are cytosolic detectors of pathogen-associated molecular patterns (PAMPs). Nod1 is a three-domain protein, consisting of a caspase activation and recruitment domain (CARD), a nucleotide-binding oligomerization domain (NOD), and a leucine-rich repeat domain (LRR). The binding of PAMPs to the LRR results in the activation of signaling through homophilic CARD-CARD interactions. Several CARD structures have been determined, including a recent NMR structure of Nod1 CARD. In contrast to the reported NMR structure, the crystal structure reported here is a dimer, where the sixth helix is swapped between two monomers. While the overall structure is very similar to the known CARD structures, this is the first report of a homodimeric CARD structure. The ability of the CARD to exist in monomeric and dimeric forms suggests another level of regulation in the activation of NLR proteins.
Organizational Affiliation:
Department of Biochemistry, Roy J. and Lucille A. Carver College of Medicine, The University of Iowa, Iowa City, IA, USA.